BACKGROUND AND AIMS
Prostate cancer (PCa) is one of the most common malignancies in men and a major cause of cancer-related death worldwide.1 PCa has a heterogeneous clinical course, with some cases exhibiting low-risk disease, while others exhibit aggressive phenotypes that quickly develop metastasis and treatment resistance. While androgen deprivation therapy is the standard treatment for metastatic cases, castration-resistant PCa is common.2 The BRCA1, BRCA2, and RAD51 genes, which are involved in the repair of DNA double-strand breaks, play a critical role in maintaining genomic stability through homologous recombination.3 Mutations or changes in expression in these genes are associated with poor prognosis and treatment resistance in many cancers, including PCa.4 The aim of this study was to determine BRCA1, BRCA2, and RAD51 gene expression levels in patients with non-metastatic PCa and to evaluate their association with clinicopathological parameters such as biochemical recurrence (BCR), Gleason score, and age.5
METHODS
Fifty patients with non-metastatic PCa who underwent radical prostatectomy between January 2020–March 2023, as well as 20 healthy controls, were included. RNA isolated from peripheral blood was converted to complementary DNA, and BRCA1, BRCA2, and RAD51 expression was measured by quantitative reverse transcription PCR using the GAPDH reference gene. Statistical analyses used parametric and non-parametric tests, Pearson correlation, and receiver operating characteristic analysis (p<0.05 was significant).
RESULTS
BRCA1 and RAD51 expressions were decreased, and BRCA2 was increased in patients with PCa compared to controls; the decrease in BRCA1 was significant (p<0.001; Table 1 and Figure 1).
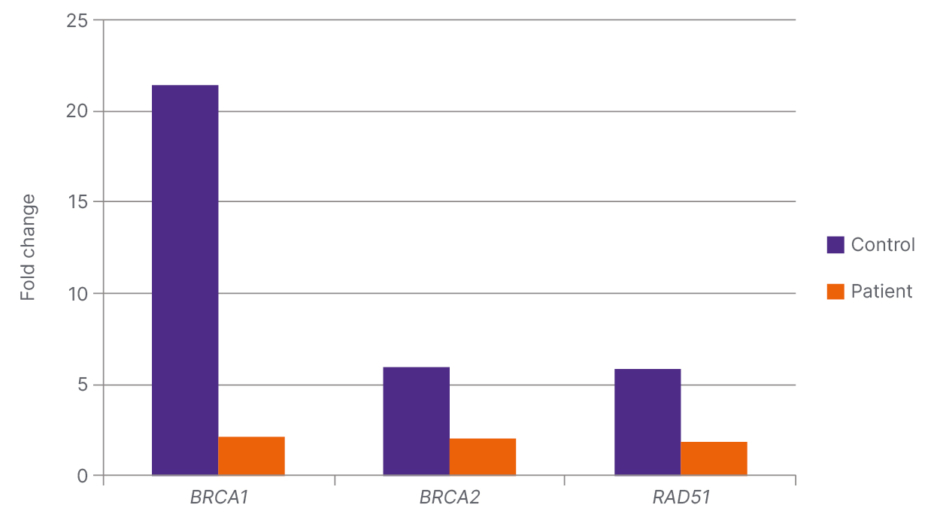
Figure 1: Fold change ratio and significance values of the genes examined in the patient (N=50) and control (N=20) groups.
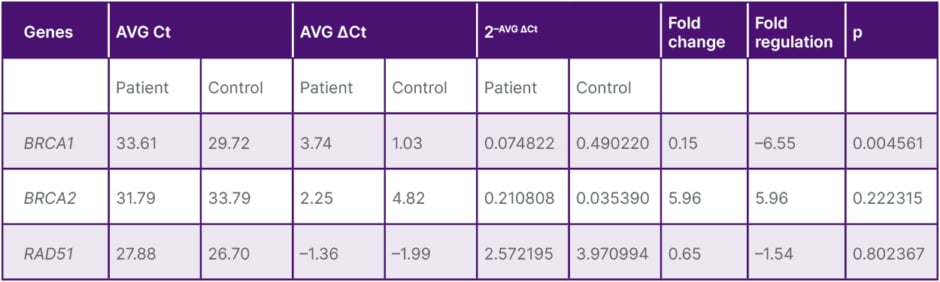
Table 1: Mean BRCA1, BRCA2, and RAD51 expression and evaluation in the patient and control groups.
p<0.05 was considered significant.
AVG: average; Ct: cycle threshold.
BRCA1 and RAD51 were increased in patients with BCR, with the BRCA1 increase demonstrating high significance (p<0.001; Table 2 and Figure 2). In patients with low Gleason scores (6–7), expression of all three genes was found to be elevated, with only the RAD51 increase being significant (p=0.019). In patients aged 60 years and older, BRCA1, BRCA2, and RAD51 levels were significantly lower (p=0.012, p<0.001, and p<0.001, respectively). A positive correlation was found between BRCA1 and RAD51 (r=0.46; p<0.01). Receiver operating characteristic analysis indicated that the combination of BRCA1 and RAD51 exhibited a significant trend in predicting BCR.
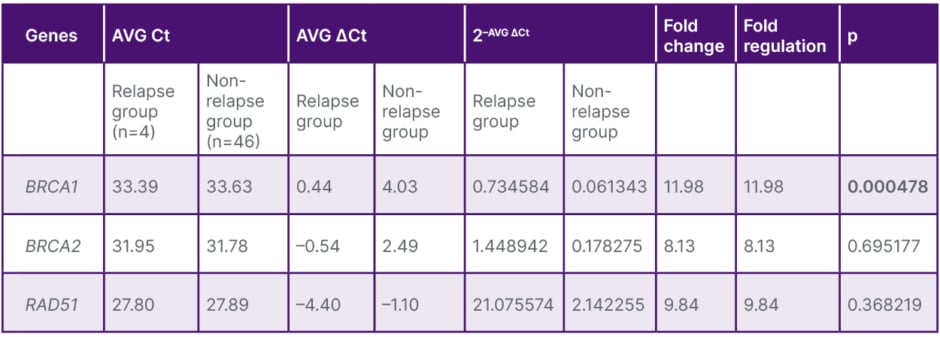
Table 2: Mean BRCA1, BRCA2, and RAD51 expression and assessment in patients with and without relapse.
The bolded p value indicates that it is significant.
AVG: average; Ct: cycle threshold.
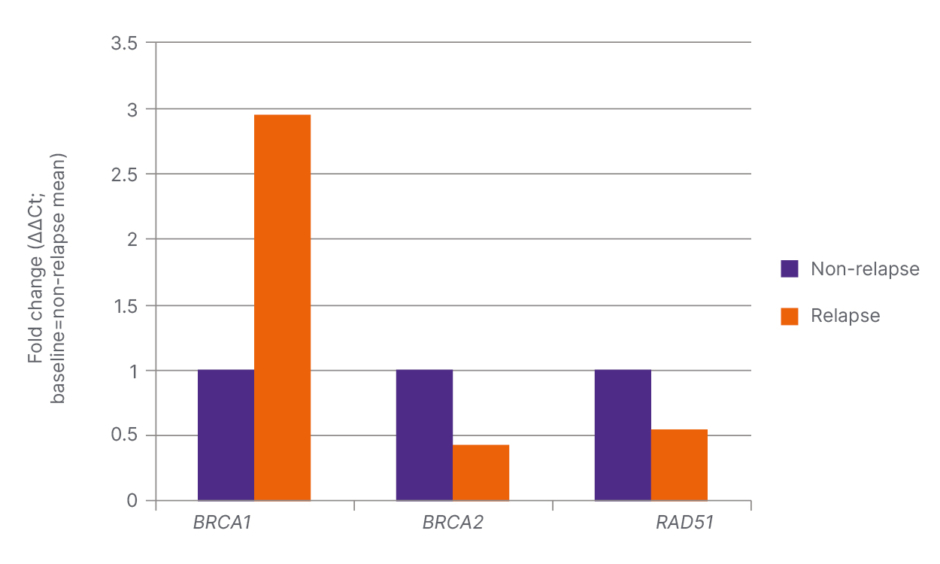
Figure 2: Fold change rates and significance values of BRCA1, BRCA2, and RAD51 genes in patient groups with and without relapse.
Ct: cycle threshold.
DISCUSSION
The increase in BRCA1 and RAD51 in patients with BCR suggests that these genes may play a role in tumour aggressiveness and recurrence risk.6,7 The high expression of RAD51 in low-grade tumours suggests that DNA repair capacity may be more effective at an early stage, while the differential expression pattern of BRCA2 may reflect its unique function in the homologous recombination mechanism.6 The literature suggests that BRCA2 mutations increase the risk of PCa by two-to-four-fold and are associated with aggressive histology and higher mortality.8,9 This study is one of the few to demonstrate the association of BRCA1 and RAD51 with age, tumour grade, and BCR in non-metastatic PCa in the same cohort.
CONCLUSION
BRCA1, BRCA2, and RAD51 genes exhibit clinically significant expression changes in non-metastatic PCa, with a strong association with age, Gleason score, and BCR. In particular, the increase in BRCA1 and RAD51 in BCR cases, and the decrease in all three genes with age, support the potential of these genes as diagnostic and prognostic biomarkers. Studies with larger series will provide valuable information for genetic screening strategies and personalised treatments.