Chairpeople: Uwe Platzbecker¹
Speakers: Uwe Platzbecker, Raphael Itzykson,² David Valcárcel³
1. University Hospital Leipzig, Leipzig, Germany
2. Hopital Saint Louis, Paris, France
3. Hospital Vall d’Hebron, Barcelona, Spain
Disclosure: Dr Platzbecker has received consulting fees and research support from Amgen, Celgene, Janssen, Merck, and Novartis. Dr Itzykson has received research funding from Bristol Myers Squibb (Celgene). Dr Valcárcel has received honoraria, expenses, consulting fees, and funded research from Celgene, Novartis, Jazz Pharmaceuticals, Janssen, Amgen, Astellas, MSD, Sanofi, and Sobi.
Acknowledgements: Writing assistance was provided by Dr Julia Duffey, Manifold Medical Communications, Matlock, UK.
Support: This content is sponsored by Novartis.
Citation: EMJ Hematol. 2021;9[Suppl 2]:2-10.
Meeting Summary
This symposium took place virtually at the European School of Haematology (ESH) 7th Translational Research e-Conference: Myelodysplastic Syndromes (MDS). Dr Itzykson began the symposium by explaining the role of precision medicine in the management of patients with MDS and how genomics is used to refine prognosis, improve prognostic accuracy, and assist in treatment decision-making. He then went on to detail how machine learning and other biological techniques can be used to optimise treatment and patient outcomes. Dr Platzbecker discussed the role of immune and inflammatory microenvironments in the pathophysiology of MDS and how research into this field enables the identification of new therapeutic targets, with a focus on checkpoint modulation. Finally, Dr Valcárcel discussed the rationale for targeting new pathways in MDS and how increased understanding of these pathways can aid the development of new therapies for the treatment of patients with higher-risk MDS.
Precision Medicine: Genomics and Beyond
Doctor Raphael Itzykson
Precision medicine is the tailoring of treatment according to the individual characteristics of a patient.1 This approach improves outcomes by taking into account both patient and disease heterogeneity.1 While the landscape of driver mutations in patients with MDS is well established,2-4 genomics is now being used to refine diagnosis,5 improve prognostic accuracy,2 and enable the selection of the best available treatment.4
Prognostic and therapeutic applications can be optimised by analysing certain mutations. For example, SF311-mutated MDS are a distinct diagnostic entity with a distinct clinical presentation, morphological footprint, and a specific co-mutation pattern.6 Multiple investigations have been performed, which assessed individual mutations to refine prognostic stratification.7 The presence of certain gene lesions have an impact on prognosis;7 furthermore, prognosis can be refined by assessing the ploidy (multiple) allelic inactivation of a mutation8 or a single allelic mutation.3,9 The size of the mutated clone may also have prognostic significance. The presence, frequency, and ploidy of the mutated gene may also impact prognosis.
MDS are oligoclonal or polyclonal diseases. The clonal hierarchies in MDS have prognostic relevance.3,10,11 Conventional prognostic scoring systems (e.g., the Revised International Prognostic Scoring System [IPSS-R] for MDS) categorise patients into groups with five possible outcomes; however, in real-world clinical practice, patients with MDS display a diverse range of outcomes. One approach to counteract intrapatient genetic diversity and interindividual heterogeneity is to collect a large data set and apply machine learning approaches to deliver personalised predictions. The era of personalised predictions using machine learning in MDS is just beginning.
Machine learning was initially proposed for patients with acute myeloid leukaemia (AML) with the development of a multistate description using a knowledge bank of 1,540 patients with AML.12 This approach was found to better identify patients suitable for allogeneic stem cell transplantation compared with genetic classifiers.13,14 Machine learning could be used in patients with MDS to guide clinical decision-making. Nazha et al.15 pioneered this technique16 by developing the ‘recommender system’ algorithm.17 This tool identifies mutation association rules and predicts responses to hypomethylating agents (HMA) by considering the specific genetic make-up of each patient.15 They also found that a random forest algorithm could predict the outcome of allogeneic stem cell transplantations better than MDS or bone marrow transplantation prognostic tools.15
Other biological techniques, beyond genomics, can be used to optimise treatment and patient outcomes. These techniques are required as not all drugs have genetic biomarkers; genotype or phenotype resistance to targeted therapies may be present before treatment initiation, or several therapeutic options for one mutation may exist.18 Functional tools have been developed to complement genomics and to refine therapeutic allocations.19 A diverse range of functional assays are available, including enzymatic assays, two- or three-dimensional culture, or cytometry assays. However, physicians should be mindful of what can be undertaken in daily clinical practice; therefore, this presentation focussed on flow cytometry and two-dimensional culture. As therapies used in patients with high-risk MDS, such as HMA, rely on specific properties of MDS cells, flow cytometry assays can guide initial treatment decisions; e.g., mitochondrial priming of selected MDS or AML cell populations can refine genetic risk stratification by incubating patients’ cells with strong apoptotic inducer peptides and computing the area under the curve of mitochondrial release of cytochrome C.20 Using this method, unprimed or primed mitochondria can be identified.20 Improved outcomes were observed in patients with primed mitochondria when treated with intensive chemotherapy.20 Functional classification of patients’ blast cells has improved the genetic classification of AML by grouping patients into favourable or intermediate chemosensitive groups.20
Current limitations of ex vivo drug screening include the reliance on artificial cell culture conditions and the overall viability of the cells used in the assay. This method also analyses drugs across a broad range of concentrations that may not be clinically relevant. To overcome these limitations, a niche-like ex vivo high-throughput (NEXT) drug screening advance platform has been developed to refine treatment decision-making in patients with secondary AML or high-risk MDS.21-24 With this platform, researchers were able to demonstrate that the tool was clinically and genetically relevant, as the platform was able to predict the depth of remission in terms of minimal residual disease (MRD) in NPM1-mutated AML treated with 7+3 chemotherapy.21 The platform also showed that patients with cognate mutations had better responses to targeted inhibitors, such as crenolanib for patients with FLT3-internal tandem duplication (FLT3-ITD), or ivosidenib for patients with an IDH1 mutation.21
The importance of precision tools was demonstrated by a study that assessed 24 drugs to find the optimal sensitiser in 48 patients with AML. The investigation found that venetoclax was the optimal sensitiser in 58% (n=24) of patients, and while the majority of patients would have benefited from venetoclax, 40% of patients would have better outcomes from other personalised drug regimens.21
In conclusion, artificial intelligence is the next step in personalised treatment selection and prognosis for patients with MDS. Further investigation of functional assays in patients with high-risk MDS will also facilitate tailored therapeutic decision-making.
The Immune and Inflammatory Microenvironments: Identifying Therapeutic Targets
Doctor Uwe Platzbecker
MDS is a heterogeneous disease with complex biology and pathophysiology that result in variable patient outcomes. This is depicted by the IPSS-R25 in which a large range in outcomes is seen in patients receiving supportive care only (range: <1 year to >8 years); this variability means that some patients do not require treatment. Furthermore, a universal therapeutic agent is not available;26 however, several new therapies are being evaluated in preclinical or clinical trials that target different pathways (Figure 1).26
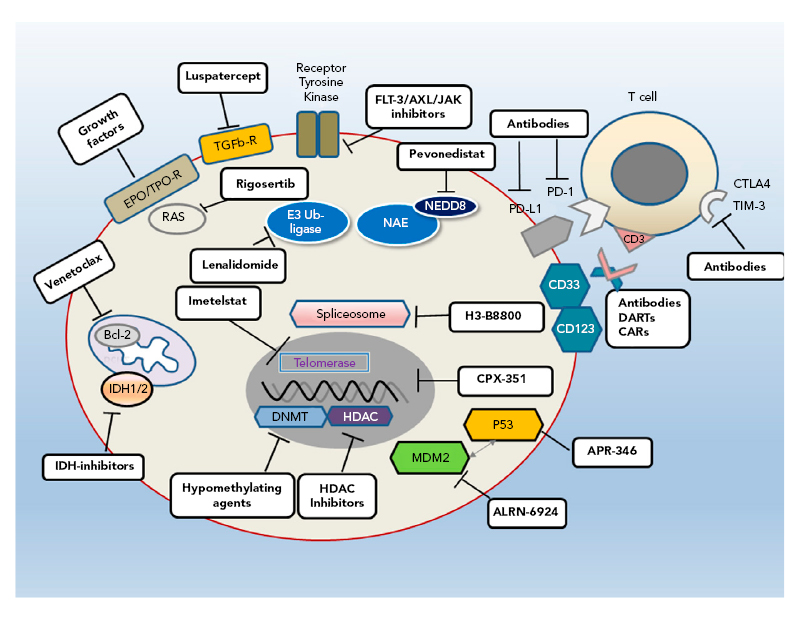
Figure 1: Different pathways provide multiple targets for developmental therapies for myelodysplastic syndromes.
Bcl-2: B-cell lymphoma 2; CAR: chimeric antigen receptor; CTLA4: cytotoxic T-lymphocyte-associated protein 4; DART: dual-affinity e-targeting antibody; DNMT: DNA methyltransferase; EPO: erythropoietin; FLT-3: FMS-like tyrosine kinase 3; HDAC: histone deacetylase; IDH: isocitrate dehydrogenase; JAK: janus kinase; MDM2: murine double minute 2; NAE: NEED8-activating enzyme; PD-1: programmed cell death protein 1; PD-L1: programmed cell death ligand 1; R: receptor; TGFb-R: transforming growth factor β receptor; TIM-3: T-cell Ig and mucin domain-3; TPO: thrombopoietin.
Reproduced with permission from Platzbecker.26
The MDS landscape is entering an era of personalised medicine for the treatment of lower-risk and higher-risk patients with MDS. Assessment of alterations in the immune system and ‘immunome’ are standard practice; however, it is not yet clear whether these alterations are implicated in the disease.27 In the early stages of MDS, inflammation driven by IL-1β and other inflammatory cytokines (inflammasomes) may be very important and contribute to the phenotype of the disease.27 In higher-risk patients, an expansion of regulatory T cells and myeloid-derived suppressor cells are observed, which may result in genetic instability and immune evasion.27
Currently, treatment options are limited to a few agents, including the recently approved luspatercept for low-risk patients with ring sideroblast phenotype. Several other agents are in development that are targeted to specific patient characteristics, including TP-53 and Bcl-2. Inflammasome activation and immune suppressive therapy are also interesting options.
Activation of the immune system is a result of a close crosstalk between the haematopoietic stem and progenitor cells and the mesenchymal stem cells in the osteohaematopoietic niche, occurring deep in the bone marrow.28 Preclinical research has identified several options for targeting inflammasome activation. Potential treatments under investigation include IL-1 receptor-associated kinase inhibitors, anti-CD33 therapies, and treatments targeted towards IL-1β or anti-IL-18.29,30
Immune checkpoints have been studied in MDS, and upregulation of programmed cell death protein 1 and programmed cell death ligand 1 on blast cells and the stromal compartment has been observed.30 In patients with higher-risk MDS failing HMA therapy, upregulation of programmed cell death protein 1 on T lymphocytes and programmed cell death ligand 1 on blast cells was reported.31 However, the differences between these findings and those of the control cohorts were moderate;31 this could explain why early clinical trials with immune checkpoint inhibitor monotherapy (ipilimumab) in patients with HMA-refractory MDS and following allogeneic stem cell transplant have shown disappointing outcomes.32,33
For example, a complete response (CR) or partial response was not reported in any patients with HMA-refractory MDS, and 20.7% of patients experienced an immune-related adverse event (AE) following ipilimumab therapy (AE ≥Grade 2).32,33 In post-allogeneic stem cell transplant patients with myeloid malignancies treated with ipilimumab, 23% reported a CR and 9% a partial response; however, 14% of patients experienced dose-limiting graft versus host disease.32,33 Data from randomised studies evaluating first-line treatment with HMA and checkpoint inhibitors in patients with MDS and AML have been disappointing, with no significant differences reported between combination therapy and HMA alone.34
The T-cell Ig domain and mucin domain-containing molecule 3 (TIM-3) protein is a novel target, inhibitors of which are under investigation. TIM-3 differs from traditional T-cell checkpoints as it is expressed on leukaemic and blast cells and on immune effector cells, where it is a marker for dysfunction/exhaustion.35-39 Inhibition of TIM-3 causes a dual effect on both the immune system and leukaemic cells. Furthermore, TIM-3 is highly overexpressed on AML stem cells but is not expressed in normal haematopoietic stem cells. TIM-3 is thought to be associated with disease severity in patients with MDS.39,40 The TIM-3/Gal-9 interaction is physiologically important to regulate leukaemic stem cell (LSC) self-renewal and promote clonal selection of the stem cell, which is mediated by NFκB and β-catenin, contributing to leukaemic progression in myeloid malignancies.41 By inhibiting this autocrine loop, the self-renewal of early leukaemic haematopoietic stem progenitor cells may be negatively affected.41
Sabatolimab is an investigational high-affinity, humanised IgG4 monoclonal antibody.40,42-44 It is a potential, first-in-class, immunotherapeutic agent, which may restore immune function by dual targeting overexpressed TIM-3 on immune effector cells, LSC, and blasts.40,42-44 Further proposed antileukaemic mechanisms include the promotion of the M1 phenotype on macrophages and the antibody-dependent cellular phagocytosis in FcγRI-expressing myeloid cells and macrophages, enhancing the phagocytic uptake of LSC.45 Sabatolimab may also inhibit the TIM-3/Gal-9 autocrine feedback loop and LSC self-renewal.45
In summary, inflammatory signalling drives the pathogenesis of MDS and new therapeutics may be developed by targeting the immune response. Promising targets include TIM-3, which has a role in leukaemic transformation and disease progression; blockade of TIM-3 by sabatolimab may restore antileukaemic immune function while also targeting LSC and blast cells.36,37,39,41-43,46,47 Therefore, there is a strong rationale for investigating sabatolimab in the management of patients with higher-risk MDS and AML.
New Therapies in the Treatment of High-Risk Myelodysplastic Syndromes
Doctor David Valcárcel
HMA48 continue to be the standard of care for patients with high-risk MDS, as Phase III trials have found that different combinations of agents with different modes of action, such as lenalidomide, vorinostat, and panobinostat in combination with azacitidine (AZA), were not associated with improved survival compared with AZA monotherapy.49 Haematopoietic stem cell transplantation is the only curative option but is limited to patients aged ≤75 years. Furthermore, several real-world studies from national registries have reported lower overall survival (OS) rates in patients receiving AZA compared to those observed in clinical trials.50-52
There are several areas where the management of patients with high-risk MDS could be improved, including AE, quality of life, treatment delivery, haematological response and transfusion independence, CR and the depth of response, OS, and event-free survival. Several novel agents are in development for patients with MDS (Figure 2). These include drugs that target specific mutations (TP-53), those that target new pathways and interfere with immune checkpoints (TIM-3), and novel formulations of classical drugs. Dr Valcárcel detailed some of these drugs, presenting recent trial data. Discussing sabatolimab, he underlined its dual activity targeting both immune effector cells and LSC. In a Phase Ib clinical trial, sabatolimab in combination with HMA (AZA or decitabine) had a favourable tolerability profile, including low rates of immune-related AE, comparable with HMA alone.53 High response rates were reported for both high-risk and very-high-risk patients with MDS receiving sabatolimab. The overall response rate (ORR) was high in the combination therapy cohort for very-high-risk/high-risk patients with MDS (64.1%), those with newly diagnosed AML (41.0%), and patients with chronic myelomonocytic leukaemia (54.5%).53 The median time to response was 2 months, with 83.9% maintenance of response at 6 months (95% confidence interval [CI]: 66.7–100.0%).53 The estimated 12-month progression-free survival was 51.9% (95% CI: 33.6–70.3%) and several patients proceeded to transplant (n=9/39; 23.1%).
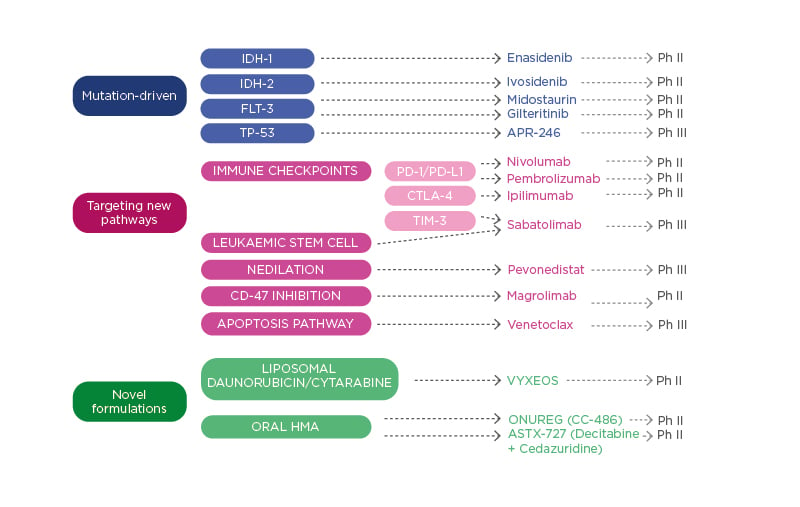
Figure 2: Therapies in development for patients with myelodysplastic syndromes.
HMA: hypomethylating agents; Ph II: Phase II trial; Ph III: Phase III trial; TIM-3: T-cell Ig and mucin domain-3.
Pevonedistat is a first-in-class NEDD8 inhibitor that interferes with DNA replication, the cell cycle, and the NFκB pathway to induce apoptosis and cell death.54 Clinical trials assessed pevonedistat in combination with AZA versus AZA alone in patients with high-risk MDS (n=67) and found no increase in toxicity.55 The median AZA dose intensity was 98% in both treatment arms, and the median number of cycles was 13.5 in the pevonedistat with AZA group versus 10.0 in the AZA group.55 The Phase II investigation (n=59) found that ORR was higher in the pevonedistat with AZA arm (79%) versus the AZA arm (57%; p=0.065).55 The CR rate was also higher in the combination arm (52%) versus the monotherapy arm (27%; p=0.05).
Median duration of response was longer for patients in the pevonedistat with AZA cohort (34.6 months; 95% CI: 11.53–34.60) than for participants in the AZA cohort (11.53 months; 95% CI: 12.02, not evaluable). Trends to improved event-free survival and OS were observed, but the cohorts were not powered to detect statistically significant differences in these outcomes.55
TP-53 mutations are common in patients with MDS (20%) and present physicians with management challenges.10 Several drugs target TP-53, with APR-246 being the most advanced. APR-246 converts into methylene quinuclidinone and leads to conformational change in mutated TP-53, increasing wild-type protein, restoring function, and allowing the cell to undergo apoptosis.56 APR-246 has been assessed in 55 patients with MDS, AML, and chronic myelomonocytic leukaemia in Phase Ib/II trials where APR-246 was compared with AZA.57 Whilst the safety profile is not as favourable as pevonedistat or sabatolimab due to neurological toxicities, clinical trial findings showed a high ORR rate of 88% and a CRR of 61%. Furthermore, 52% of patients progressed to transplant.57 Publication of final results of Phase III studies is awaited to see whether they support Phase II findings.
In summary, over the next few years results from diagnostic panels will be available and some patients will be able to receive personalised and tailored combination therapies. After initial treatments, physicians will still need to evaluate whether patients are candidates for treatment with post-transplant therapy to reduce relapse rates. If a patient is not a candidate for transplant, then responses should be assessed as soon as possible to maintain or adjust therapy for optimal outcomes.
Panel Discussion
Question and Answer
How are daily clinical data transferred to the databases for computational analysis? How can we assure the quality of the data in the generation of precision-based models? How is this achieved in your clinical practice?
Dr Itzykson explained that currently, precision medicine approaches have only used available databases, with data coded by investigators based on real-life data and specific response criteria; however, response criteria are imperfect, and researchers are constantly seeking to improve them. It is a challenge to determine whether the optimal approach is intermediate data processed by humans or the use of daily raw health data sets. Both approaches have advantages and disadvantages: there are clinical confounders that can blur the specific assessments of response criteria, yet patients may feel that the capture of all data might be intrusive. Dr Platzbecker concluded that human intelligence is important and should always be the backbone of analyses.
Do you believe that venetoclax is going to be the next standard of care in high-risk MDS?
Dr Valcárcel outlined that there are some positive data on venetoclax in combination with HMA, but it is uncertain if this combination will be appropriate for all high-risk patients or if other drugs such as pevonedistat in combination with HMA will be used. Several factors that should be taken into consideration are the mechanism of action and the toxicities associated with each agent. Dr Platzbecker stated that it might be possible to combine two or three therapies in the future.
For patients with high-risk MDS, how important will measuring MRD become, and will sequencing of therapy and maintenance therapy be integrated into clinical practice in the future?
Dr Valcárcelexplained that both measuring MRD and sequential therapy to evaluate responsiveness may be important. Currently, sequential therapy is challenging as the therapies are not available. Dr Itzykson explained that MRD makes the fundamental assumption that you have “good” cells that are invaded by a compartment of “bad” cells and the purpose of treatment is to eradicate the bad cells. However, in patients with MDS there is a continuous spectrum of disease; MRD may not be the optimal surrogate endpoint at all points on the spectrum, and other interesting endpoints may be relevant.
Can we eradicate or modulate haematopoiesis in the future?
Dr Itzykson outlined that this is more challenging than originally thought. In most patients, clones will not develop into frank myelodysplasia, so targeting clonal haematopoiesis routes of leukaemia may not be the best option. Dr Platzbecker concluded that the optimal approach might be to repopulate the bone marrow after inflammation without further haematopoiesis progression.







