Abstract
Peritoneal dialysis (PD) is a cost-effective, home-based treatment option for patients with end-stage renal disease; however, PD is declining in many countries. A major reason for this is peritonitis, which commonly leads to technique failure and has led to negative perceptions of PD by clinicians and patients. To restore confidence in PD, better diagnostics are required to enable appropriate treatment to be started earlier; this needs to be coupled with improved understanding of the biology of peritonitis. Advances in culture-independent microbiological methods, in particular the use of bacterial flow cytometry and immune fingerprinting techniques, can enable organism detection and antimicrobial susceptibility testing to be performed in as little as 3 hours after samples are received. At the same time, improved understanding of peritoneal mesothelial cell responses to infection is providing insights into pathways that may be targeted to dampen deleterious elementsof the host immune response, promote healing, and preserve membrane function.
INTRODUCTION
Worldwide, >2.5 million people have end-stage renal disease and are receiving renal replacement therapy.1 Over 250,000 patients with end-stage renal disease are treated with peritoneal dialysis (PD) worldwide,2 which equates to ˜10% of all dialysis patients, but this figure can be as high as 60–70% in some countries where a PD first strategy is employed.3,4 PD is cost-effective, offers a better quality of life compared to haemodialysis, and, in some settings, may be the only available treatment option. The annual global growth rate of PD is estimated to be ˜8%, which is higher than for haemodialysis (˜6–7%).2 However, the proportion of dialysis patients treated with PD is declining in developed countries, despite PD being associated with superior survival in the first few years, better quality of life, and lower treatment costs; this is, therefore, of great concern.5-8
PD uses a catheter placed into the abdomen, with instillation of dialysis solutions of varying composition to enable fluid and toxin removal; as a result, a major complication of PD is the development of a peritonitis infection. Peritonitis is the single largest cause of patients failing on PD; approximately half of all PD technique failures are due to peritonitis, with peritoneal infection also strongly associated with mortality.9 Fear of peritonitis is a major reason for patients and clinicians not choosing PD.10 Gram-positive cocci, such as Staphylococcus epidermidis and other staphylococcal species, are the most frequent cause of PD-associated peritonitis worldwide, with Gram-negative organisms accounting for 20–25% of cases and fungal infections ~4%.11,12 Despite the use of broad spectrum antibiotics when patients present with peritonitis, many develop relapsing or recurrent life-threatening infections. Even when treatment is successful, deleterious changes may occur in peritoneal membrane function, which ultimately lead to inadequate solute or fluid removal and technique failure.
The treatment of, and outcomes from, peritonitis are highly variable between countries and even within medical centres in the same country. This is despite the publication of treatment guidelines.13 This suggests that despite decades of research and clinical experience, there remains concerns regarding guideline content and that there is a lack of consensus about the management of peritonitis.14-16 A major barrier to improving the treatment of patients with peritonitis is the use of traditional, culture-based diagnostic microbiology to confirm the presence of infection. These techniques are slow, with cultures usually taking 1–3 days to become positive, which can cause delays in diagnosis. In addition, many organisms are either difficult or impossible to culture,17 with reported culture-negative rates of up to 20% in some centres.11 As a result, clinicians commence empirical antimicrobial therapy based on historical profiles and published guidelines rather than patient-specific laboratory evidence. Even when cultures are positive, definitive antimicrobial susceptibility results that enable the tailoring of antibiotic treatment to the most effective regimen often require a further 1–3 days. This delay likely contributes to the increased mortality and morbidity of patients on PD and the emergence of drug-resistant microbes.18,19 Reducing the time taken for clinicians to receive results that guide effective therapeutic decision-making is therefore critical to achieving better outcomes for patients.
A better understanding of the molecular pathways that control infection (susceptibility, initiation, severity, recovery, and/or relapse) should enable their manipulation to improve outcomes and reduce peritoneal membrane damage. New advances in culture-independent diagnostic methods and knowledge of mesothelial cells and peritoneal responses to infection provide hope that much-needed improvements in peritonitis outcomes are in sight.
ADVANCING CULTURE-INDEPENDENT MICROBIOLOGY
A number of culture-independent laboratory methods are now available that promise faster, more sensitive, and more specific aetiological diagnoses across a broad spectrum of pathogen and specimen types.20 Examples include nucleic acid-based approaches to detect bacteria-specific DNA or RNA, and protein-based assays, such as matrix assisted laser desorption/ionisation time-of-flight (MALDI-TOF) mass spectrometry (MS), which identifies organism-unique protein signatures.21,22 These techniques are relatively rapid, leading to faster reporting times and the detection of organisms that may be difficult to culture.23 Each approach, however, has significant limitations. They do not distinguish viable from nonviable organisms, the number of defined genetic targets for nucleic acid-based detection are currently limited, and the interpretation of results can be complicated in polymicrobial infections contaminating species and/or the presence of commensal bacteria.23,24 In addition, even if specific resistance genes are detected by polymerase chain reaction (PCR), expression of these genes may vary and multiple genes may be required to yield functional resistance in vivo.25,26
To date, neither bacterial nucleic acid nor protein-based detection techniques are routinely used for analysis of samples from patients with suspected PD peritonitis.27 Major limitations of these techniques for the analysis of PD samples are the high bacterial concentrations required for adequate sensitivity during protein detection28,29 and the poor accuracy of nucleic acid detection, which has unacceptably high false negative rates, thought to be due to the presence of inhibitors in dialysate (Figure 1).30 Bacterially derived DNA fragments in PD effluent show some value as a prognostic marker for relapsing peritonitis episodes, but as the presence of bacterial DNA does not directly correlate with live organisms capable of causing infection, the clinical applicability is limited.31 23S ribosomal (r)RNA PCR and sequencing has been applied to the problem; however, the authors concluded that this method was best reserved as an adjunctive tool to traditional culture techniques given the lack of specificity when applied as a diagnostic test.32 While not an exhaustive set of examples, this illustrates the complexity faced when attempting to use nucleic acid-based technology in a sample as complicated as PD effluent.
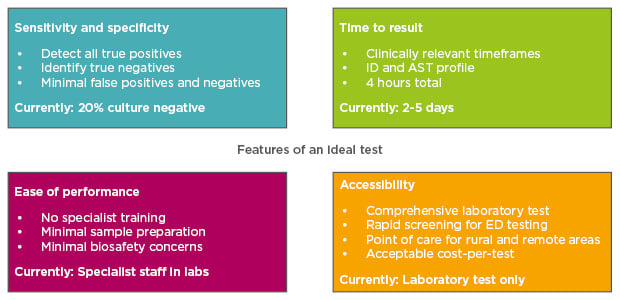
Figure 1: Features of an ideal diagnostic test for kidney disease.
AST: antimicrobial susceptibility testing; ED: emergency department; ID: identification of infecting organism.
ADVANCES IN FLOW CYTOMETRY ENABLE DIRECT VISUALISATION OF BACTERIA AND FUNGI
While flow cytometry (FC) has been extensively used to detect eukaryotic cells, the smaller size of bacteria coupled with variability in cell wall structures has limited its use for bacterial detection.33 However, recent advances in hardware design, including the use of acoustic-focussing technology (Figure 2A), have greatly improved the effective resolution capable for small particles. Reliable detection of biological particles as small as 250 nm is now considered routine.34 Coupling this technology with DNA-intercalating and protein-binding fluorescent dyes improves resolution further and makes accurate identification of bacteria or fungi directly from clinical samples possible (Figure 2B). The quantitative nature of particle characterisation by FC also permits direct enumeration of bacterial counts in dialysate providing information on inoculum dose, which may be an important feature in influencing clinical outcomes.
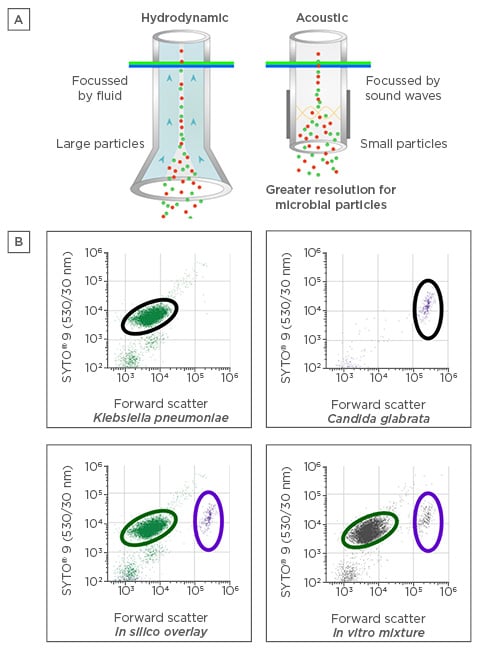
Figure 2: A) Acoustic versus standard hydrodynamic focussing for flow cytometry. B) Clear separation between bacterial and fungal pathogens in peritoneal dialysis dialysate as assessed by acoustic flow cytometry.
SYTO®, Thermo Fisher Scientific, Waltham, Massachusetts, USA.
One unique aspect of this approach is the separation of bacterial detection from identification of their species (or Gram type), which may prove challenging for clinicians and microbiologists who have traditionally decided upon treatment options based on this knowledge. While this has implications for epidemiological data collection (if a traditional culture is not also conducted),35 this approach can provide answers where traditional culture techniques have failed, for example in cases of culture-negative peritonitis, or where recent exposure to antimicrobial agents might impact culture. Furthermore, sample preparation for this technique can require as little as 25 minutes of manual handling, followed by 10 minutes for data acquisition and processing, representing a potential gain of >18 hours compared to traditional culture-based methods.36
CULTURE-INDEPENDENT ANTIMICROBIAL SUSCEPTIBILITY TESTING
Most antimicrobial agents used in the treatment of PD peritonitis have bacterial (or fungal) cell lysis as their final mechanism of action. As FC detection of this mechanism works by identifying cell shape, size, and wall integrity, the effects of antibiotics on cells can be analysed and antimicrobial susceptibility profiles determined ex vivo in samples. This method, termed flow-assisted antimicrobial sensitivity testing (FAST), has demonstrated a strong positive correlation (r2: 0.81; p<0.0001) with current international standard culture-dependent methods, and has been demonstrated to be useful for common PD pathogens across a range of relevant antimicrobials (Figure 3).36 Extensive development work is ongoing to assess the application of this technology to detect bacteria directly from PD effluent and determine the cost-effectiveness of this approach and its performance compared to standard culture-based assays. As a guide, currently a 96-well plate for antimicrobial susceptibility profiling with 12 minimum inhibitory concentrations being assessed costs <$100, including consumables and technician time.
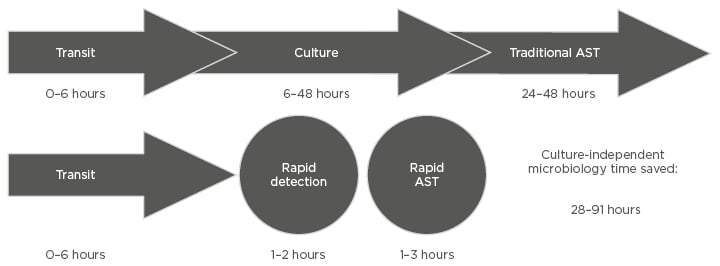
Figure 3: Comparison of timeframes for culture-dependent and culture-independent diagnostic methods to provide results in.
AST: antimicrobial susceptibility testing.
While improved diagnostic tools are clearly essential and have demonstrated promise in controlled situations with small cohorts, the majority of patients who develop peritonitis receive effective antimicrobial therapy. In these cases, peritonitis resolves within 7–10 days and most patients are managed in an outpatient setting. However, epidemiological data demonstrate clear differences in outcomes depending on the class of organism causing peritonitis, with Gram-negative and fungal species generally associated with higher morbidity and mortality.37 Even in successful respondents, the impact of peritonitis on the long-term durability of PD is significant, with a high incidence of membrane dysfunction, leading to inadequate fluid and solute removal and technique failure.38,39 To alter these outcomes, detection must be improved to provide more rapid and targeted treatment, and, even more importantly, understanding of the host responses to different types of infection needs to be improved. Only by elucidating all the factors can the clinical course of PD peritonitis be better understood (resolution versus relapse, benign versus severe). This will allow the ability to develop new or modify existing treatments to reduce morbidity and mortality, and to preserve and protect the peritoneal membrane so that patients can continue on PD for longer.
IMMUNE FINGERPRINTING
As opposed to detecting the infecting micro-organisms directly, an alternative approach has been to study pathogen-specific host responses instead, which has particular advantages in settings where organism numbers are below the detection limit of current tests. One novel approach to faster identification of bacterial peritonitis is to analyse the nature of the host response elicited by infection. This technique, known as ‘immune fingerprinting’, utilises the fact that humans have evolved specific mechanisms to detect and respond to invading micro-organisms that the species have encountered throughout millions of years of evolution.24,40 These pattern recognition receptors and their signalling pathways define the initial host response to microbial infection, and systematic analysis of samples from patients with PD peritonitis has demonstrated that a unique signature can be identified that accurately distinguishes Gram-negative, Gram-positive, and culture-negative peritonitis at presentation, which are important distinctions when deciding upon the most appropriate treatment.24,41 These fingerprints can be rapidly determined through measurement of secreted proteins (present in PD fluid) and may be suitable for the development of point-of-care testing. Although this technique does not provide information about antimicrobial susceptibility profiles, which still rely on standard cultures being performed, it is highly complementary to organism-based approaches as it integrates the host responses into a comprehensive view of local infection and may offer additional insights into virulence and immunopathology. Studies assessing the accuracy and efficacy of immune fingerprinting and culture-independent approaches in comparison to standard traditional culture techniques are ongoing, and these studies will also provide important information on the real-world cost of these assays.
UNDERSTANDING PERITONEAL IMMUNITY AND ITS PROGNOSTIC SIGNIFICANCE
The central players in the initial host response to infection are the local immune and non-immune cells that survey the peritoneal cavity in the steady state and respond rapidly to injury and infection. While the role of peritoneal macrophages in the local inflammatory response has been established,42,43 the contribution of mesothelial cells, a single cell monolayer that lines the visceral and parietal surfaces of organs within the abdominal and chest cavities,44 to peritoneal immunity remains less well understood. These cells are highly metabolically active, have phagocytic properties, and produce numerous cytokines,45,46 which recruit other immune cells and initiate repair processes following resolution of infection.47,48
Few studies have directly assessed how different bacteria affect mesothelial cells, despite clear evidence that this influences clinical outcomes.49,50 Recent work has shown that mesothelial cells vary widely in their responses to S. epidermidis isolates cultured from patients with PD peritonitis,51 indicating that unique characteristics of the specific bacterial isolates are responsible for the different patterns of gene expression observed. The variation in individual peritoneal inflammatory responses seen in peritonitis episodes caused by the same species clearly indicates that the quality and magnitude of the activation process varies significantly. This may be related to many factors, including the individual’s immune competence, genetic variants that influence responses,52 the growth environment, the time between the start of the infection and presentation to hospital and commencement of treatment, and the virulence of the infecting organism. To date, this has not been examined in detail but may be of great significance in understanding the relationship between patient inflammatory responses and long-term outcomes. For example, recent analysis of gδ T cell numbers and their activation by bacteria has been linked to technique failure and mortality in PD patients.24,41
Significant genetic variation between isolates of the same bacteria is known to occur, with many bacterial species capable of exchanging segments of DNA, which can contain genes that confer virulence or fitness advantages to the isolate under specific environmental conditions (e.g., iron acquisition systems, antibiotic resistance genes, protein secretions systems, invasins, adhesins, and toxins).53-55 These genomic islands have been identified in all of the major bacterial species responsible for PD peritonitis,11 including S. epidermidis,56,57 Staphylococcus aureus,58 Streptococcus spp.,59 Enterococcus spp.,60 Klebsiella spp.,61 and Pseudomonas spp.62
Studies to assess the impact of individual bacteria and virulence factors on mesothelial cell activation, and the role that different dialysate solutions may play in modulating their expression and the host response, will help identify why particular types of infections promote long-term membrane damage and has the potential to discover pathways that may be therapeutic targets. These studies will require systematic analysis of mesothelial cell responses to pure isolates of bacteria cultured in spent dialysate from patients and correlation of these results to hard clinical endpoints. Given the complexity of the biological processes involved in PD peritonitis, such analyses will benefit greatly from the emerging power of systems biology-based approaches and the integration of mathematical modelling in experimental and observational studies.41
CONCLUSION
Currently, the assessment of infection has not advanced substantially since the times of Anton van Leeuwenhoek, Robert Koch, and Louis Pasteur, with reliance on microscopy and culture of patient specimens to confirm the diagnosis and guide appropriate treatment. This outdated approach has left the medical community with high incidences of culture-negative peritonitis in some units and relatively slow turnaround times. As a consequence, clinicians continue to rely on broad-spectrum empirical therapies, exposing patients to potentially unnecessary treatments with potential side effects, including the development and spread of antimicrobial resistance and high rates of long-term technique failure. Advances in laboratory technology over the past decade, particularly with culture-independent microbiology, are showing promise. However, despite repeated calls in international guidelines and research initiatives for translation of these new techniques to the clinical setting, progress has been slow. Even with the promise of improved and more rapid diagnostics, our understanding of host–microbe interactions in the peritoneal cavity remains poor. This needs to be a focus of ongoing research if we are to understand how infection-driven inflammation contributes to poor outcomes and to use this for diagnostic and prognostic gain. This knowledge will guide more rapid and effective interventions and lead to an increase in patient and clinician confidence in PD as a therapy through achieving better membrane preservation and long-term outcomes.