BACKGROUND AND AIMS
COPD is a highly prevalent condition, but it often remains underdiagnosed due to its nonspecific clinical presentation and reliance on technically demanding spirometry.1 Blood-based biomarkers could help in identifying individuals at risk and offer insights into disease pathophysiology.2 This study aimed to identify metabolic alterations associated with COPD and to develop a biological signature potentially useful for screening purposes.
METHODS
A case-control study design was used, including 182 fasting plasma samples from 91 patients with COPD and 91 controls (individuals who either formerly smoked or currently smoke, without airflow limitation), drawn from the BIOMEPOC3 and EARLY-COPD4 multicenter Spanish cohorts. Patients underwent extensive clinical characterization to match both groups for age, sex, BMI, and smoking history. Metabolomic profiling was performed using a combination of semi-targeted and untargeted liquid chromatography-tandem mass spectrometry, providing broad metabolite coverage and improving identification accuracy. Only metabolites detected in ≥80% of samples were included in the analysis. Data processing and statistical analyses were carried out using MetaboAnalyst 6.0.5 Differentially abundant metabolites were identified through comparative analysis adjusted for multiple comparisons (false discovery rate <0.1). These were further examined using multiple regression analysis controlling for confounding variables, including age, sex, BMI, and plasma cotinine levels (as a marker of recent smoking). Pathway enrichment analyses were conducted using the Human Metabolome Database and the Kyoto Encyclopedia of Genes and Genomes database. For biomarker discovery, support vector machine models were applied with 100 Monte Carlo cross-validation iterations to derive a minimal 10-metabolite panel, excluding xenobiotics to enhance endogenous biological relevance.
RESULTS
Both the COPD and control groups included adults predominantly in their 40s to 60s, with a balanced sex distribution (52% male) and a mean BMI within the overweight range (26 kg/m2). Patients with COPD exhibited impaired respiratory function characterized by airflow obstruction and reduced diffusing capacity for carbon monoxide, with most classified (using the Global Initiative for Chronic Obstructive Lung Disease [GOLD] classification) as GOLD 2, followed by GOLD 3 and GOLD 1.6,7
Out of 360 quantifiable metabolites, 74 differed significantly between COPD and controls, with 56 remaining significant after adjustment for confounders. The main endogenous changes were related to lipid metabolism, including both over- and under-representation of specific fatty acids and acylcarnitines, as well as alterations in amino acid and carbohydrate metabolism. Notably, the largest group of differentially abundant metabolites consisted of xenobiotics. These non-endogenous compounds likely reflect environmental or microbiota-derived exposures, and were therefore excluded from the final diagnostic model to preserve biological relevance.6,7
The final 10-metabolite endogenous panel comprised of N-methylglutamate, diethanolamine, and gluconic acid (increased), as well as leucic acid, palmitic acid, 14-methylhexadecanoic acid, 2-hydroxymyristic acid, glyceric acid, 2-aminonicotinic acid, and urocanate (decreased). It achieved high discriminatory power (area under the curve: 0.916; 90.1% sensitivity; 89% specificity; Figure 1). Pathway analysis highlighted disruptions in fatty acid β-oxidation, amino acid catabolism, and the pentose phosphate pathway.6,7
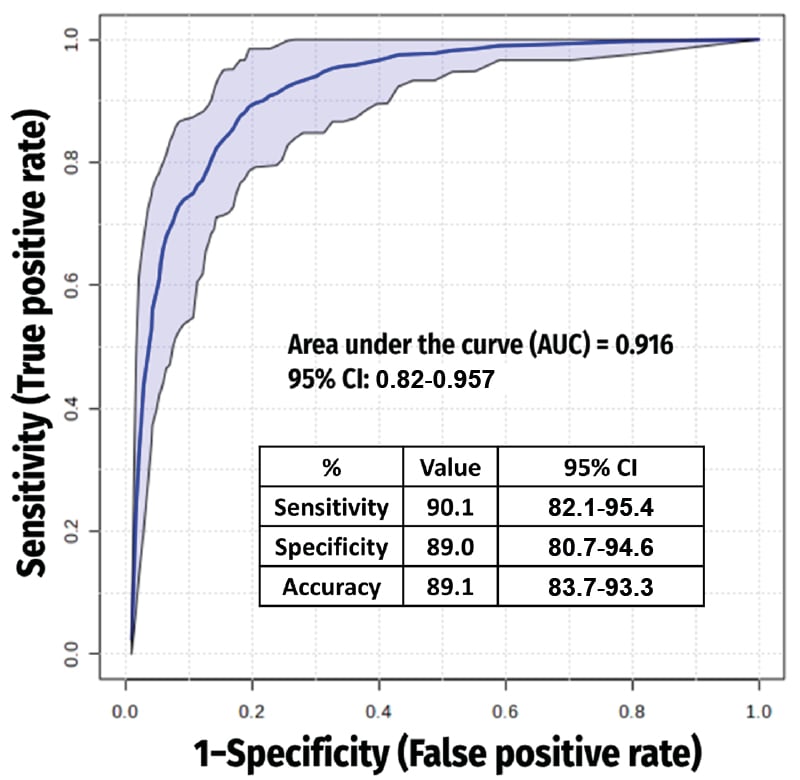
Figure 1: Receiver operating characteristic curve from the best model (which included 10 metabolites).
CONCLUSION
The exclusive use of endogenous metabolites reinforces both biological plausibility and clinical translatability. The identified metabolic profile provides novel insights into energy metabolism, inflammation, and redox balance in young and old COPD. This may aid in screening individuals at high-risk. Nonetheless, further mechanistic research and long-term external validation are required to confirm causality. Despite this, the proposed signature marks a step forward in precision diagnostics for COPD.
This study identifies a xenobiotic-free 10-metabolite plasma panel that allows for high-accuracy COPD diagnosis and sheds light on the disease’s underlying metabolic disturbances. Plasma metabolomics reveals a distinct, physiologically relevant COPD profile. Future studies should validate its clinical utility and investigate therapeutic strategies targeting altered metabolic pathways.