Abstract
The integration of AI into healthcare has marked a transformative era, with machine learning (ML) playing a pivotal role in reshaping the landscape of interstitial lung disease (ILD) management. ML models excel in analysing complex datasets, such as medical imaging and electronic health records, offering unprecedented advancements in the diagnosis, prognosis, and treatment of ILDs. These models have demonstrated superior accuracy compared to traditional methods, particularly in diagnosing idiopathic pulmonary fibrosis, where delays in diagnosis significantly impact patient outcomes. Early and precise diagnosis through ML-driven tools allows for the timely initiation of therapy, which is crucial for improving prognosis and extending patients’ quality of life. Despite the challenges in data quality and model interpretability, the future of ML in pulmonary healthcare is promising, with continued advancements poised to enhance patient management and outcomes. This article aims to examine the transformative potential of ML in the management of ILD.
Key Points
1. Interstitial lung diseases (ILD) cause significant morbidity, yet diagnosis is often delayed. AI offers potential to improve early detection, prognostication, and treatment selection, addressing major unmet needs in ILD care.2. This narrative review synthesised 26 primary peer-reviewed studies applying machine learning to ILD, covering diagnostic imaging, biomarker discovery, and prognostic modelling, with comparisons to human readers and evaluation of emerging AI tools.
3. Machine learning can match or surpass expert performance in ILD diagnosis, predict progression, and identify novel biomarkers, but widespread clinical adoption requires prospective validation, interpretability, and integration into real-world workflows.
INTRODUCTION
In recent years, the convergence of AI and healthcare has ushered in a transformative era marked by unprecedented advancements.1 At the heart of this revolution lies machine learning (ML), a subset of AI that empowers systems to learn from data, identify patterns, and make decisions with minimal human intervention.1 ML, broadly defined, involves algorithms that enable computers to learn from and analyse large volumes of data.1 These algorithms improve their performance through iterative processes, making predictions or decisions based on historical data.1 In healthcare, ML models offer the potential to enhance diagnostic accuracy, prognostic insights, and treatment planning.
Traditional diagnostic methods often rely on subjective interpretation of symptoms and diagnostic tests, which can be influenced by human error and variability. ML algorithms, however, can analyse complex datasets, including medical imaging, genetic information, and electronic health records (EHR), to identify subtle patterns that may elude human practitioners. This capability can lead to earlier and more accurate diagnoses, ultimately improving patient outcomes.
Additionally, ML enables personalised prognostic modelling by integrating diverse data such as patient demographics, lifestyle factors, and clinical history. This allows for individualised risk assessments and predictions about disease progression and treatment response, helping to optimise clinical decision-making and potentially prevent adverse outcomes.2
Interstitial lung diseases (ILD) represent a heterogeneous group of approximately 200 pulmonary disorders, characterised by varying degrees of inflammation and fibrosis affecting the lung interstitium.3 Accurate diagnosis often requires a multidisciplinary approach, combining clinical, radiological, and pathological assessments, given overlapping imaging patterns and heterogeneous presentations.4 The traditional diagnostic process for ILD is often lengthy and invasive. Patients may undergo multiple evaluations, and in complex cases, a surgical lung biopsy is sometimes required to establish a definitive diagnosis. Even with multidisciplinary discussions, misdiagnoses or significant diagnostic delays are common. Notably, emerging AI systems may facilitate the detection of pulmonary fibrosis even before overt clinical or radiological manifestations appear. For instance, a new AI-driven screening tool was able to predict pulmonary fibrosis up to 4 years before a conventional diagnosis (area under the receiver operating characteristic curve [AUROC]: ~0.84 at 4 years),5 underscoring the potential of ML in identifying ILD at a pre-fibrotic stage, when early intervention could be most beneficial.
Idiopathic pulmonary fibrosis (IPF) is often diagnosed late, with a median delay of 2.1 years, largely due to misdiagnosis and delays at multiple healthcare levels, including general practitioners and community hospitals.6 This delay is particularly concerning, given that early initiation of therapy in IPF is associated with better outcomes and slower disease progression.7
In summary, ML holds transformative potential in the realm of ILDs, offering advancements in diagnosis, prognosis, and treatment.8 As the technology continues to evolve, it is poised to enhance patient management and outcomes, marking a significant leap forward in the quest for more effective and personalised healthcare. As highlighted by Barnes et al.,9 ML represents a ‘new frontier’ in radiology for ILD, offering a shift from subjective pattern recognition toward reproducible, high-throughput analysis.9
METHODOLOGY
Literature Search Strategy
To explore the impact of ML techniques in ILD, a comprehensive search of relevant literature was conducted. The authors searched the following databases from 1999–20 June 2025:
- PubMed;
- IEEE Xplore;
- Google Scholar; and
- Cochrane Library.
Boolean operators were used to combine relevant terms, such as:
- ‘interstitial lung disease’ or ‘idiopathic pulmonary fibrosis’ or ILD or IPF; and
- ‘artificial intelligence’ or ‘machine learning’ or ‘deep learning’ or ‘neural networks’ or ‘radiomics’ or ‘computer-assisted diagnosis’.
Filters were applied to restrict results to English-language, peer-reviewed, human-subject studies. Titles and abstracts were screened for relevance, followed by full-text review. Reference lists of key studies were also manually screened for additional sources. A Preferred Reporting Items for Systematic Reviews and Meta-Analyses (PRISMA)-style flowchart (Figure 1) illustrates the selection process.
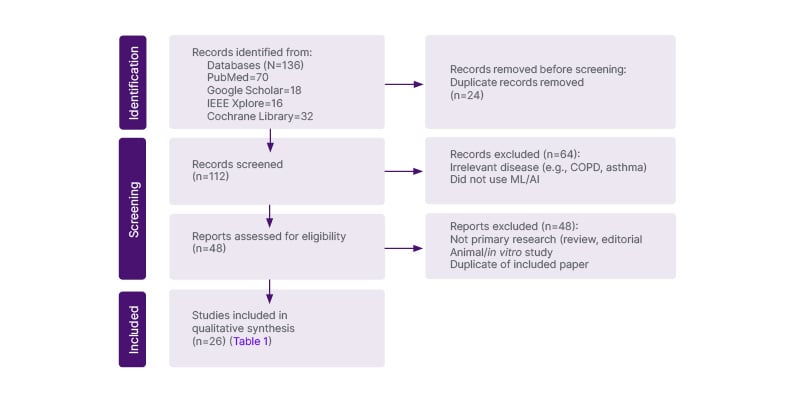
Figure 1: PRISMA flow diagram of literature selection for a review of machine learning applications in interstitial lung disease.
ML: machine learning.
Studies included in this review met the following criteria:
- Involved human subjects diagnosed with any subtype of ILD;
- Applied AI or ML techniques for diagnostic, prognostic, or biomarkerdiscovery purposes; and
- Reported specific outcomes, such as model performance metrics (e.g., AUROC, sensitivity, specificity), diagnostic accuracy, or clinical utility.
A total of 26 primary studies were included in the authors’ results, each applying ML techniques to human subjects with ILD and reporting relevant diagnostic, prognostic, or biomarker outcomes. An additional set of systematic reviews and expert perspectives was referenced in the discussion to contextualise findings, but was not included in the formal study count.
Exclusion criteria included studies not directly related to ML applications in ILDs, studies focused solely on non-human subjects, studies lacking sufficient detail on methodology or results, and non-peer-reviewed material, including editorials, commentaries, conference abstracts, and technical white papers.
Data Synthesis and Analysis
A narrative synthesis approach was used due to the substantial heterogeneity among included studies in terms of ML models, data types, ILD subtypes, and outcome measures. Given the early-stage and exploratory nature of AI applications in ILD, with limited standardisation across methodologies, a narrative framework allowed for meaningful comparison, critical appraisal, and integration of diverse findings that would not be suitable for quantitative meta-analysis.
A total of 26 studies were included and analysed for this review, focusing on the application of ML in ILD. A summary of the results is shown below in Table 1.
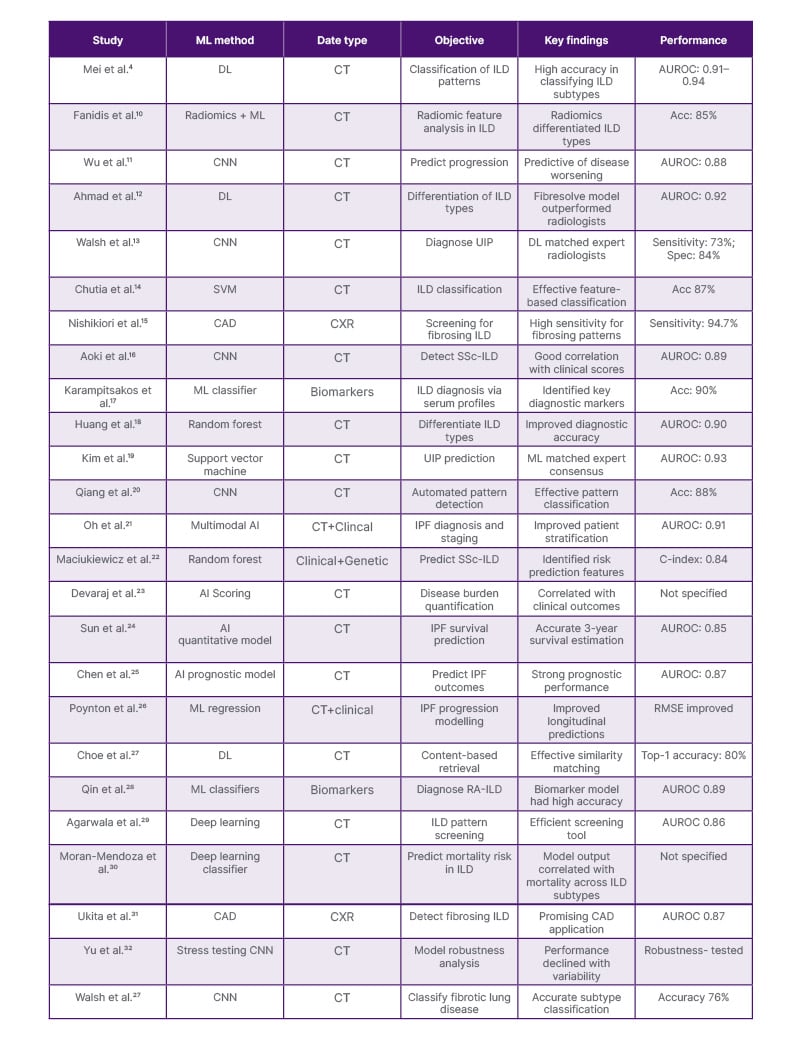
Table 1: Summary of key studies on machine learning applications in interstitial lung disease.
Acc: accuracy; AUROC: area under the receiver operating characteristic curve; C-index: concordance index; CAD: computer-aided detection; CNN: convolutional neural network; CXR: chest X-ray; DL: deep learning; ILD: interstitial lung disease; IPF: idiopathic pulmonary fibrosis; ML: machine learning; RA-ILD: rheumatoid arthritis–associated interstitial lung disease; RMSE: root mean square error; Spec: specificity; SS-ILD: systemic sclerosis–associated interstitial lung disease; SVM: support vector machine; UIP: usual interstitial pneumonia.
DISCUSSION
The application of ML in ILD represents a significant advancement in healthcare, offering promising improvements in diagnosis, prognosis, and treatment. As ML technologies continue to evolve, they provide increasingly sophisticated tools for analysing complex medical data, potentially addressing some of the longstanding challenges in ILD management. A pivotal aspect of these advancements is the comparison of ML models with traditional diagnostic methods, including human readers, and the impact of these comparisons on clinical practice. Mekov et al.34 offered an early overview, outlining how AI tools could bridge radiologic and clinical domains by supporting differential diagnosis and care planning in respiratory medicine, while Chan and Auffermann35 emphasised the potential of AI to unify multimodal imaging in diffuse lung diseases.
Machine Learning in the Diagnosis of Interstitial Lung Disease
Radiological evaluation of ILD, particularly the identification and characterisation of pulmonary fibrosis, presents persistent challenges, even for experienced thoracic radiologists.36 A key difficulty lies in detecting honeycombing, a defining feature of usual interstitial pneumonia (UIP), which is central to diagnosis but subject to high interobserver variability and diagnostic uncertainty.36 This diagnostic ambiguity is especially pronounced in patients who do not meet criteria for a definitive UIP pattern.37
Chang et al.38 addressed this clinical gap by training an ML classifier on CT scans labelled with pathology- and clinical-supported diagnoses, intentionally excluding cases with clear UIP to focus on patients who are classified as ‘grey zone’, where radiologic interpretation alone may be insufficient.38 Such models are especially valuable in real-world practice, where diagnostic confidence varies and multidisciplinary discussion is often required.
Building on this, Castillo-Saldana et al.39 applied quantitative CT metrics to distinguish fibrotic ILD from emphysema, a common diagnostic dilemma. By leveraging densitometric and histogram-based features, their model captured subtle structural differences not readily appreciated by visual inspection, suggesting a role for quantitative imaging in phenotyping patients with overlapping clinical or radiographic features.39 Complementing this, Ukita et al.31 developed a deep learning (DL)–based computer-aided detection system to identify fibrosing ILD on plain chest radiographs. Though the study did not directly compare ILD to emphysema, its use in a broad screening context highlights the potential of computer-aided detection tools to navigate diagnostic ambiguity and improve early identification.31
Convolutional neural networks (CNN)are particularly effective in medical imaging due to their structure and functionality.40 They start by representing an input image as a grid of numbers, with each number indicating the brightness of a pixel. CNNs use small squares called filters that slide across the image, performing a mathematical operation known as convolution to highlight specific features like edges or colours.40 Following this, pooling reduces the image’s size by keeping only the most important parts, allowing the network to focus on key features while enhancing processing speed.40 CNNs consist of multiple layers, with each layer recognising increasingly complex patterns, from basic edges in early layers to complete objects in later ones. Ultimately, the network classifies the image, identifying it as, for example, a cat or a dog, based on the features it has learned. This process enables CNNs to analyse and interpret a wide variety of images effectively. One of the key advantages of CNNs is their translation invariance, meaning they can recognise objects regardless of their position in the image. Beyond medical imaging, CNNs are widely used in real-world applications, including facialrecognition and autonomous driving.40
In a pivotal study by Mei et al.,4 CNN and vision transformer models were evaluated for both ILD subtype classification and survival prediction.4 Using CT scans and clinical data, the joint CNN–multilayer perceptron model achieved an AUROC of 0.94, significantly outperforming a panel of seven human readers, whose combined AUROC was 0.88. The panel included radiologists and pulmonologists with varying experience levels, all of whom were provided identical CT scans and clinical metadata. The model also demonstrated higher sensitivity (90%) and specificity (87%) for diagnosing UIP compared to readers (sensitivity: 80%; specificity: 83%).4 These findings show the potential of ML to enhance diagnostic precision in ILD, particularly in complex or borderline cases. Still, the single-centre nature of this study warrants cautious interpretation until external validation is achieved.
In an early study from India, Agarwala et al.29 developed a DL algorithm to detect ILD patterns on high-resolution CT, achieving an AUROC of 0.91.29 This work is particularly notable for demonstrating the scalability and adaptability of AI models across diverse healthcare systems, including resource-limited environments.
Ahmad et al.12 developed Fibresolve, an ML tool designed to identify IPF from other ILDs using thin-slice CT imaging.12 Notably, the algorithm outperformed clinical panels in cases with atypical UIP patterns that often require surgical biopsy for definitive diagnosis. Among patients who did not meet imaging criteria for IPF but had ≤3 mm CT slices, Fibresolve achieved a diagnostic yield of 53.1% and a specificity of 85.9%. These figures are particularly meaningful, considering that traditional diagnostic pathways for such cases are often prolonged and invasive. By reducing the median time to diagnosis (213 days), Fibresolve could meaningfully expedite care and reduce the need for invasive procedures. The system has since received FDA approval, further supporting its potential utility in clinical practice.
Walsh et al.33 developed a DLalgorithm using 1,157 anonymised, high-resolution CT scans to classify fibrotic lung disease.33 The algorithm achieved an accuracy of 76.4%, surpassing 66% of 91 thoracic radiologists, whose median accuracy was 70.7%. Additionally, the algorithm showed good interobserver agreement (weighted kappa [κw ]=0.69), exceeding 62% of the radiologists (κw=0.67), and offered near-instantaneous diagnoses, taking only 2.31 seconds to evaluate 150 four-slice montages. This rapid and reproducible performance highlights the efficiency and reliability of ML algorithms compared to human readers,33 but we must bear in mind that real-world performance may vary, and such systems would require rigorous external validation.As Yu et al.32 point out, real-world performance may diverge from training data benchmarks. Their study retrospectively evaluated DL models for IPF diagnosis and found variability in performance when applied to different institutions and CT acquisition protocols, emphasising the importance of cross-site robustness testing before clinical deployment.32
A notable development in ML for ILD diagnosis was achieved by researchers at Sapporo Medical University Hospital, Japan, who created a DL model for detecting chronic fibrosing ILDs using chest radiographs.15 This model, which is the first to employ chest radiographs instead of CT scans, achieved an impressive area under the curve (AUC) of 0.979, with a sensitivity of 0.896 and specificity of 1.000. This performance is comparable to that of experienced radiologists and pulmonologists, demonstrating the model’s potential as a valuable diagnostic tool.15 In the realm of histopathology, Fukuoka et al.41 conducted a large international study demonstrating that AI could help standardise histopathologic diagnoses of UIP by reducing interobserver variability among expert pathologists, establishing a potential reference framework for future diagnostic tools.41 Complementing this, Chung et al.42 evaluated a genomic classifier capable of identifying UIP even in patients lacking classic high-resolution CT patterns, reinforcing the value of AI-driven molecular diagnostics in complex or ambiguous cases.42
It is also worth mentioning that, although most current AI applications in ILD are geared toward recognition of fibrotic disease patterns, there is increasing recognition that identifying pre-fibrotic interstitial abnormalities, such as interstitial lung abnormalities or early non-specific interstitial pneumonia, can improve clinical outcomes by enabling earlier intervention.13 Incorporating AI into radiologic and histopathologic pipelines may enhance pattern recognition of subtle pre-fibrotic changes and support multidisciplinary team decision-making before irreversible damage occurs. AI’s application in identifying pre-fibrotic conditions remains an underexplored but crucial frontier. Early detection, even before irreversible fibrosis sets in, could substantially improve long-term outcomes and reduce the need for invasive diagnostics.
Finally, content-based image retrieval systems are emerging as novel AI tools with both diagnostic and educational value. Choe et al.27 developed a DL-based content-based image retrieval system that retrieves visually similar annotated CT scans to assist with ILD subtype recognition, achieving an AUROC of 0.922 for distinguishing UIP from nonspecific interstitial pneumonia.27 These tools may enhance radiologists’ confidence, reduce ambiguity in borderline cases, and promote standardisation across institutions.
Machine Learning in Biomarker Discovery for IdiopathicPulmonary Fibrosis
While imaging remains the cornerstone of ILD diagnosis, biomarker discovery through ML is an increasingly active and promising frontier. These approaches aim to augment diagnostic accuracy, stratify risk, and ultimately tailor treatment by extracting patterns from high-dimensional molecular data, spanning transcriptomics, proteomics, and gene expression profiling.
A notable example is the work by Kim et al.,19 who applied ML to high-dimensional transcriptional data to classify UIP versus non-UIP patterns in ILD.19 Their model demonstrated high diagnostic accuracy, supporting the potential of molecular classifiers as adjuncts to radiologic and histopathologic assessment. This early, yet pivotal, study laid the groundwork for multi-omic ML models, bridging the gap between molecular pathology and clinical phenotyping in ILD. Building on this, Huang et al.18 extended the scope to plasma proteomics, applying ML to quantify over 1,300 proteins from patients with ILD and controls.18 Their model achieved near-perfect discrimination (AUROC: 0.99 for ILD versus control; AUROC: 0.90 for IPF versus non-IPF), suggesting that proteomic signatures may soon complement imaging in classifying ILD subtypes. Importantly, this study highlights how proteomics could enable earlier and less invasive diagnosis if validated in external cohorts.
Fanidis et al.10 employed the eXtreme gradient boosting ML algorithm on gene expression data to explore potential molecular signatures associated with pulmonary fibrosis.10 The model achieved an encouraging accuracy (range: 0.85–0.95) and identified several candidate genes, including IL13Rα2 and PAPSS2, with possible roles in fibrotic pathways. IL13Rα2 is a key receptor that IL-13 uses to induce fibrosis, and its signalling is crucial for the production of TGF-β,43 a major contributor to fibrotic processes in chronic inflammatory diseases.44,45 To interpret the model’s predictions, Shapley additive explanation analysis was utilised, quantifying the contribution of each feature (gene) to the overall prediction. This methodology offers insight into model decision-making and helped to identify 76 candidate genes potentially associated with fibrosis. While these findings highlight promising avenues for further investigation, it is important to note that these biomarkers remain exploratory and have yet to undergo validation in large, prospective cohorts.
Wu et al.11 conducted a parallel study focusing on differentially expressed genes and identified four critical biomarkers: FHL2, HPCAL1, RNF182, and SLAIN1.11 These genes have demonstrated validated predictive value, particularly highlighting SLAIN1 for its potential role in informing future therapeutic strategies. Notably, FHL2 has been associated with tissue remodelling and fibrosis, further emphasising its significance within the context of IPF. Although these genes demonstrate predictive potential within retrospective datasets, their utility as diagnostic or therapeutic biomarkers also requires further clinical validation, including reproducibility across diverse populations.
Lastly, Qin et al.28 focused on rheumatoid arthritis-associated ILD, developing ML classifiers using support vector machines and random forests to detect transcriptomic signatures specific to this subset.28 Their model yielded strong diagnostic performance (AUROC: 0.89), reinforcing the idea that ML can help surface disease-specific molecular fingerprints in clinically overlapping ILD phenotypes.
Machine Learning in Interstitial Lung Disease Prognosis
In their expert review, Bendstrup et al.45 emphasised the importance of structured ILD monitoring using symptoms, spirometry, and imaging. These routine clinical touchpoints offer a natural opportunity for ML to augment care, whether by automating change detection on chest CTs or flagging subtle declines in pulmonary function tests before they cross conventional thresholds.45 Within this prognostic domain, Chutia et al.14 developed a model to predict lung function decline in IPF by analysing 1,554 forced vital capacity (FVC) records from 176 patients, along with demographic data, smoking status, and CT scans.14 Using quantile regression combined with CNNs, the model achieved a striking 92% accuracy in forecasting lung function decline, supporting ML’s potential to inform timely intervention and improve disease monitoring.
Imaging-based models have alsoshown significant promise. Chen et al.25 trained a DL algorithm topredict mortality in IPF using chest CT features, achieving high predictive accuracy and reinforcing the role of imaging biomarkers in prognosis.25 Similarly, Aoki et al.16 demonstrated that a DL-based quantification tool correlated strongly with FVC and diffusing capacityof the lungs for carbon monoxide,and achieved an AUROC of 0.78 for predicting ILD progression.16 Thesefindings highlight the utility ofquantitative CT metrics as surrogates for physiologic decline when automated via deep learning pipelines.
Expanding this work, Teramachi et al.46 developed a longitudinal DL model that incorporated clinical data and environmental exposures to predict acute exacerbations and mortality in patients with ILD.46 Walsh et al.,13 who had earlier applied DL to classify fibrotic lung disease, extended their model to predict mortality in progressive fibrotic ILD, demonstrating the broader applicability of AI-derived radiologic scores in outcome prediction.13 Likewise, Moran-Mendoza et al.30 found that their ML–derived CT classifier score correlated significantly with mortality in a real-world ILD cohort, highlighting the prognostic potential of AI beyond simple subtype classification.30
Other models have extended ML-based prognostic prediction to rare ILD phenotypes. For instance, Qiang et al.20 trained a random forest model on CT and serum biomarkers to predict rapid progression in idiopathic inflammatory myopathy-associated ILD, achieving an AUC of 0.883.20 Oh et al.21 similarly demonstrated that DL-derived fibrosis extent on CT predicted transplant-free survival independently of radiologist-assigned pattern.21 A related application of radiomics-based ML was explored by Karampitsakos et al.,17 who developed a random forest classifier trained on quantitative CT features to predict fibrotic ILD progression in survivors of COVID-19.17 Their model achieved robust predictive performance at 3 and 6 months (AUC: 0.827 and 0.851, respectively), demonstrating the adaptability of ML-based prognostic tools beyond idiopathic disease and into viral-induced ILD phenotypes. Other radiomics applications further support the utility of quantitative imaging. Chassagnon et al.37 developed an automated DL system to assess ILD severity in systemic sclerosis using CT imaging.37 Maciukiewicz et al.22 demonstrated that radiomic features from high-resolution CTs could predict FVC decline in systemic sclerosis-associated ILD using a random forest classifier.22 Sun et al.24 applied ensemble learning in connective tissue disease–associated ILD, integrating demographics, radiographic data, and pulmonary function tests to predict long-term mortality.24
Recent studies have highlighted the prognostic potential of imaging-based ML models. In a post hoc analysis of a Phase II trial, Devaraj et al.23 used the e-Lung platform to derive the weighted reticulovascular score (WRVS), which outperformed traditional metrics such as diffusing capacity of the lungs for carbon monoxide in predicting disease progression in patients with IPF over 52 weeks.23
In the study conducted by Mei et al.,4 they also developed a model for ILD prognosis.4 The study aimed to predict 3-year mortality in patients with ILD using advanced ML models: a Long Short-Term Memory model and a transformer model. Both models incorporated 165 features, 32 high-level CT features extracted from chest CT scans using a pretrained CNN model, and 18 clinical variables, such as medication history and treatment details. These features were longitudinally assessed to create dynamic models for survival prediction.4 The transformermodel consistently outperformedthe Long Short-Term Memory model, showing a 15.8% better performance.
By the third year, the transformermodel’s AUROC of 0.868 indicated strong predictive performance for 3-year mortality, signifying that the model could distinguish between patients who would survive and those who would not with high accuracy.4 The model’s negative predictive values (ranging from 89.66–94.55%) suggest that it was particularly reliable at identifying patients who would survive, minimising false negatives. The increase in sensitivity from 54.55% after 1 year to 72.73% by the third year further demonstrates that the model became more accurate in identifying patients at risk of death as more follow-up data was added. This improvement highlights the importance of continuous clinical monitoring, with the model gaining more predictive power as patient history and response to treatment accumulate over time. This suggests that longerfollow-up periods allow for more accurate prognosis and could help clinicians make more informed decisions about patient management.4
Radiomics has also shown promisein detecting subclinical progression. Poynton et al.26 applied radiomicanalysis to serial chest CTs in high-risk individuals and successfully differentiated progressive interstitial lung abnormalities from stable cases, often before overt clinical or functional decline was apparent.26 This highlights the role of radiomics in early surveillance strategies. Molecular markers may further augment prognostic models. Libra et al.47 recently proposed candidate plasma biomarkers for IPF progression using ML-based analysis, offering a glimpse into how future prognostic tools might integrate multi-omic data to personalise risk stratification.47
Finally, the Duke EMPOWER app (Duke University Health System, Durham,North Carolina, USA) exemplifies the integration of digital tools with AI to enhance patient engagement and research participation.48 By offering ILD-specific education, enabling self-screening for research studies, and collecting longitudinal data on patient outcomes and biometric measures, the app illustrates a practical use of technology in managing rare diseases.48 Its success in increasing study enrolment and promoting healthy behaviours underscores the potential for AI-driven tools to improve clinical research and patient management. This direct-to-patient approach presents a promising model for other conditions, bridging the gap between technology and personalised healthcare.
Challenges and Future Directions
Despite the promising advancements in ML for diagnosing and managing ILD, several challenges must be addressed to fully realise its potential (Figure 2). One significant challenge is the integration of ML systems with existing electronic health record systems. Seamless integration is crucial for ensuring that ML tools are easily accessible and effectively utilised in clinical practice. However, this integration often involves complex technical and regulatory hurdles, including interoperability issues and the need for robust data exchange protocols. Addressing these challenges will be essential for the widespread adoption of ML tools in healthcare.
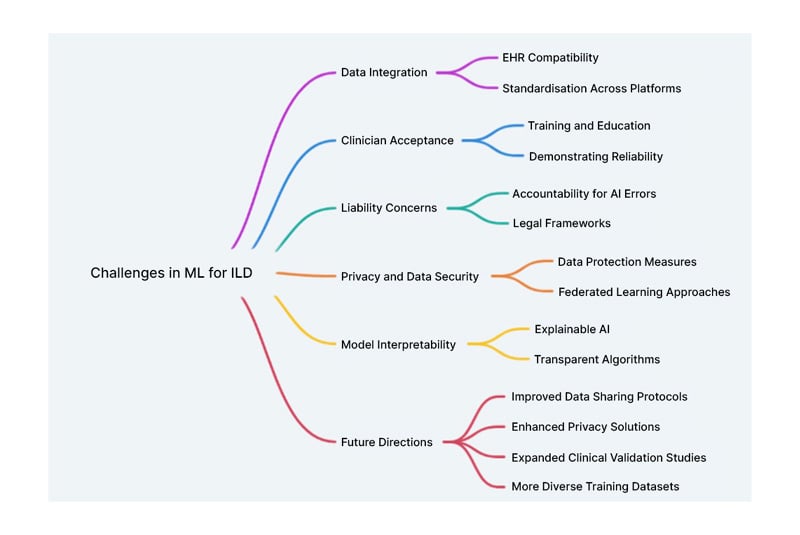
Figure 2: Challenges and future directions of machine learning in interstitial lung disease.
EHR: electronic health record; ILD: interstitial lung disease; ML: machine learning.
The generalisability of current models remains a significant limitation.49 Of the seven studies included in this review, four focus exclusively on IPF, which, though clinically significant, represents only a small subset of the more than 200 ILD subtypes. As a result, current ML models are often optimised for the recognition of fibrotic patterns associated with IPF, potentially limiting their diagnostic performance when applied to less common or non-fibrosing ILDs such as sarcoidosis, hypersensitivity pneumonitis, or connective tissue disease-associated ILD. Also, expanding model training and validation across the full ILD spectrum is essential to ensure broader clinical applicability and to prevent inequities in diagnosis and treatment. In parallel with these diagnostic innovations, Soffer et al.50 conducted a comprehensive systematic review of AI applications in ILD, highlighting the growing body of work focused on chest CT analysis.50 Their review emphasises the heterogeneity in model architectures, training datasets, and outcome definitions, which collectively pose barriers to reproducibility and clinical adoption. Importantly, they call for greater standardisation and transparency in AI development, a theme echoed throughout the authors’ review. These findings reinforce the need for interpretable and externally validated models before widespread implementation in clinical workflows.
Additionally, a challenge lies in the availability and quality of training data. ILDs are relatively rare and heterogeneous diseases, and the development of robust ML models requires large, well-annotated, and structured datasets, resources typically available only at large academic or tertiary care centres. This concentration of data introduces potential biases, as models may underperform in community settings or underserved populations where disease presentations, imaging protocols, and EHRs may differ. Furthermore, smaller centres often lack the infrastructure to collect high-resolution imaging data or comprehensive clinical annotations necessary for model development.49 Addressing this limitation will require greater collaboration across institutions, federated learning approaches, and efforts to democratise access to high-quality ILD datasets.
Another obstacle is acceptance of AI among clinicians, hospitals, and patients. Many healthcare professionals may be hesitant to rely on AI due to concerns about its reliability and the potential for reduced human oversight.51,52 Building trust in AI systems will require demonstrating their efficacy through rigorous validation studies and providing adequate training for users.51,52 Additionally, patient acceptance of AI-driven diagnostics will depend on transparency about how these tools work and how patient data is handled.
Liability concerns also pose a significant challenge.53 Determining accountability when AI systems make mistakes is complex. If an AI system provides an incorrect diagnosis or treatment recommendation, it raises questions about who is responsible: the developers, the healthcare providers, or the institutions using the technology. Clear guidelines and legal frameworks will be necessary to address these issues and ensure that patients receive safe and effective care.53
Interpretability remains a key challenge to AI adoption in ILD, as many high-performing models, such as CNNs and transformers, operate as ‘black boxes’ with limited transparency. This lack of explainability can hinder clinician trust and complicate integration. Clinicians are understandably hesitant to act on an AI’s prediction without understanding the basis, especially in high-stakes diagnoses like ILD subtyping or prognostication.
Regulatory Considerations
To advance clinical impact, futureresearch should prioritise prospective validation and implementation studies that assess ML tools in real-world settings, measuring outcomes like diagnostic accuracy, workflow integration, and patient benefit. These studies are essential to move beyond retrospective analysis and ensure meaningful clinical adoption.Privacy concerns are another critical issue. The use of AI in healthcare involves processing vast amounts of sensitive patient data and ensuring the protection of this data; addressing potential privacy breaches is paramount.54 Strategies such as federated learning, which allows models to be trained on decentralised data without compromising privacy, offer promising solutions but require further development and validation.55
From a regulatory and ethical perspective, compliance with evolving frameworks is critical. The FDA has released guidance on Software as a Medical Device (SaMD) and adaptive AI systems, emphasising transparency, clinical validation, and post-market monitoring.53,56 ML tools for ILD must align with these standards, particularly when offering diagnostic suggestions that could influence patient care. Likewise, adherence to data privacy regulations, such as the Health Insurance Portability and Accountability Act (HIPAA) in the USA, and the General Data Protection Regulation (GDPR) in the European Union, is paramount. These frameworks mandate strict governance over patient data, informed consent, and secure data storage and transfer, especially when training models on multi-institutional or international datasets.
Finally, model deployment should emphasise ethical use, explainability, and integration into clinical workflows. Collaboration with regulatory bodies, clinicians, and patients will be key to ensuring that AI tools are safe, effective, and trusted in practice.57 AI should be viewed not as a replacement for expert clinical judgment but as an assistive tool aimed at improving diagnostic consistency and efficiency in diverse care settings.58
CONCLUSION
ML is revolutionising the diagnosis and management of ILD. From CNNs and vision transformers that have outperformed experienced human experts, to integrative models that combine imaging and clinical data, ML has demonstrated substantial potential for improving diagnostic accuracy and patient outcomes. Applications like the Fibresolve system and biomarker discovery pipelines also highlight the versatility and promise of ML in ILD care.
Although these advancements exist,real-world clinical adoption remainslimited. To bridge this gap, future research must prioritise prospective validation studies across diverse clinical settings. Such studies should assess not only diagnostic and prognostic accuracy, but also the impact of ML tools on clinical decision-making, patient outcomes, and workflow efficiency. Furthermore, model deployment studies are needed toevaluate integration with EHRs, clinician usability, and patient acceptability.
Future work should also prioritise the development of interpretable models to enhance clinician trust and transparency, external validation across multicentre and multinational cohorts to strengthen generalisability, the adoption of federated learning and secure data-sharing frameworks to address privacy concerns and expand access to high-quality ILD datasets, and early regulatory engagement to ensure alignment with standards set by the FDA, HIPAA, and GDPR.
Realising the full clinical potential of ML in ILD will require a coordinated approach that emphasises prospective evaluation, ethical implementation, and multidisciplinary collaboration. Progress in this space will depend on integration into real-world practice.