BACKGROUND
Atopic dermatitis (AD) is a multifactorial, chronic, relapsing inflammatory skin disease.1,2 Characteristics are an impaired skin barrier and an altered skin immune system, which often come along with predominant colonisation by Staphylococcus aureus.3,4 The role of fungi, i.e., the mycobiome, remains poorly investigated although patients with AD are frequently sensitised to Malassezia spp., the most abundant fungus on skin.5 Through this study, the authors aimed to improve the understanding of the skin mycobiome in AD.
METHODS
Skin swabs of 15 patients with AD (nine mild-to-moderate and six severe cases) and 11 healthy controls (HC) were taken from four skin sites (antecubital crease, glabella, vertex, and dorsal neck). To assess temporal shifts in the mycobiome, patients with AD were sampled at three time points (0, 2, and 4 weeks). HC were sampled at four time points (0, 4, 8, 12 weeks). The authors analysed relative abundance of fungal classes and species by amplicon-based next-generation sequencing of the fungal internal transcribed spacer 1 region. Next-generation sequencing data were analysed with R.6
RESULTS
The most abundant fungi at all skin sites were Malassezia spp. As shown in Figure 1, patients with severe AD tended to be more frequently colonised with non-Malassezia fungi such as saccharomycetes (predominantly Candida spp.), whereas patients with mild-to-moderate AD had similar distributions as HC. The Malassezia species with the highest prevalence were M. restricta and M. globosa with a site-dependent distribution. M. restricta was most abundant at the glabella and vertex; in contrast, M. globosa had the highest abundance antecubital and at the neck. M. restricta abundance was decreased in patients with severe AD compared with the other groups, whereas, M. globosa was overall equally abundant in the different groups. In most HC and patients with mild-to-moderate AD, the mycobiome was comparable between individuals and stable over time. In contrast, in severe AD the mycobiome was different between individuals and changed over time (data not shown).
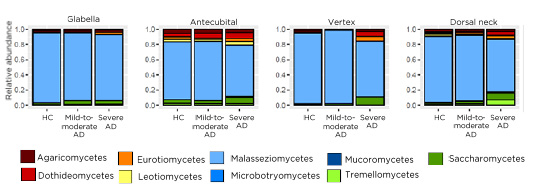
Figure 1: Merged relative abundance of fungal classes of healthy controls, patients with mild-to-moderate atopic dermatitis, and patients with severe atopic dermatitis on four different skin sites: glabella, antecubital, vertex, dorsal neck.
AD: atopic dermatitis HC: healthy controls.
CONCLUSION
Patients with severe AD had a high intra- and interpersonal species diversity. The authors speculated that the impaired skin barrier in severe AD allows colonisation with more different fungi than healthy skin. Vice versa, the altered mycobiome may cause activation of the skin immune system leading to inflammation and eczema. In the next step, the authors will correlate these results with the bacterial microbiome in the same samples.






