Abstract
Introduction: When the COVID-19 pandemic arose, samples from patients infected with SARS-CoV-2 were needed to study the new virus, and a new ‘Biobank COVID-19 Ticino’ of patients was established. The Biobank gathered biological specimens and high-quality data on the first pandemic wave to elucidate clinical presentation, natural history, response to treatment, immune response and cytokine activation, and outcomes of the disease.
Material and methods: The authors collected a full set of clinical and biological data (nasopharyngeal swab, blood, stool and urine samples together with a PAXgene [PreAnalytiX GmbH, Zurich, Switzerland]) from the patients at baseline and at scheduled follow-ups until 1 year after the infection. Patients hospitalised at Moncucco clinic in Lugano, Switzerland, who were older than 18 years and positive for a SARS-CoV-2 swab, were eligible for the Biobank.
Results: A total of 135 patients, hospitalised in Fondazione Epatocentro Ticino, Switzerland, from 25th April 2020–13th December 2021, were included. The mean age of patients was 65 years. Most participants in the COVID-19 Biobank were male (68.9%) and White (98.5%). Two patients were hospitalised in the ICU directly during the enrollment visit, while 133 patients were hospitalised in the internal medicine ward. Of the latter, 16 patients’ clinical condition worsened, and 12 required ICU admission, of whom one died. Data from this Biobank have supported four nested research projects.
Discussion and conclusion: The project of Biobank COVID-19 Ticino was created with the primary objective of collecting data and samples of patients who were hospitalised with COVID-19. During the project, the authors collected a total of 10,116 specimens. The authors’ biobank is essential because it provides high-quality, well-annotated biological samples that enable reliable research, foster medical discoveries, and accelerate the development of personalised treatments.
Key Points
1. The Biobank COVID-19 Ticino, established in 2020, collected a full set of data from 135 patients hospitalised with COVID-19.2. This represents an important dataset captured during the first wave of the pandemic. These samples remain valuable, as they have supported the development of several nested projects.
3. The future of medicine and research lies in the collection of consistent, comprehensive datasets that pair patient samples and clinical records for specific diseases. Here, patients were followed for a full year after hospitalisation, allowing researchers access to important records that extend beyond the narrow window of infection.
INTRODUCTION
When the COVID-19 pandemic reached Europe, little was known. It was mostly reported by the Chinese authorities, and therefore, healthcare professionals did not know what they were dealing with or how to treat this infection.1
In early March 2020, the authors’ region was the first in Switzerland to be confronted with rapidly increasing numbers of cases. It became clear that clinical data and biological samples from patients who were infected with SARS-CoV-2 were a very important tool for future research projects. To this end, the Fondazione Epatocentro Ticino, Swtizerland, decided to establish a new Swiss web-based cohort that would gather biological and clinical data from patients hospitalised because of COVID-19 at Clinica Luganese Moncucco in Lugano, Switzerland.2
The aim of the authors’ project was to collect high-quality data on this first phase of the pandemic and build a ‘Biobank COVID-19 Ticino’, with samples collected from patients with COVID-19 who were hospitalised at the Clinica Luganese Moncucco at different time-points. The cohort provides a platform for carrying out scientific research projects on COVID-19, and allows collaborations with reference networks.3,4
The stored samples were intended to be used to answer research questions. They are unique in the sense that they were collected during the first pandemic wave of a novel, highly contagious coronavirus interacting with an immunologically naïve population. The authors herein describe the patient population, their clinical characteristics, and the set of biological samples that they collected. They also provide a brief overview of some of the research projects done using their biobank.
METHODS OF SAMPLING
Within the Biobank data collection, after getting the written informed consent from the patients, the authors obtained biological samples suitable for genetic, serological, microbiological, and immunological studies, which have been stored in order to facilitate such studies. All patients who were not conscious at arrival were excluded because of a lack of informed consent. All enrolled patients were assigned a unique COVID-19 Biobank code, and all further information was anonymised. The authors collected clinical data with a case report form (CRF) filled in by the physician, alongside data derived from the electronic patient record. All data were collected at baseline (day of hospitalisation) and at scheduled follow-up visits, up until 1 year after the infection. Further samples were collected after any clinically relevant change, including worsening or amelioration of clinical conditions.
Worsening was considered as evolution towards pneumonia or acute respiratory distress syndrome, worsening of clinical or laboratory parameter values according to medical judgement, implementation of therapy (such as administration of tocilizumab or remdesivir), admission to the ICU, dyspnoea, and desaturation that led to intubation and mechanical ventilation or extracorporeal membrane oxygenation or death.
Amelioration was considered as extubation, no further need for oxygen therapy, and/or discharge from the ICU.
Collection of samples was performed by the trained nurses at the clinic, while labelling and storage were conducted by a dedicated clinical research unit staff. Missing data were reported in the RedCAP (REDCap Consortium, Nashville, Tennessee, USA) form for each patient and registered in the dedicated log.
In order to minimise data entry bias and errors, double-entry validation was performed by the clinical research unit members. Whenever possible, the authors aimed to obtain a complete data set of samples for each patient, considering the hospitalisation and critical conditions during the pandemic. This approach allowed them to collect the highest-quality data set possible.
Ethical Approval
The COVID-19 Biobank project was approved by the local ethics committee (Comitato Etico del Canton Ticino) and registered with the project ID 2020-00771. All patients signed the informed consent before enrolment and were assigned a unique Biobank code.
Enrolment
At enrolment, a nasal and a nasopharyngeal swab, a blood sample (three tubes of blood samples included), a sputum sample, a urine sample, and a stool sample were collected. In addition, at ICU admission, one tracheal secretion was collected and stored. A questionnaire (Appendix 1 and 2) was filled out by the physician, which focused on patients’ characteristics, vital signs, oxygen levels, early warning score, medical history, and symptoms. Laboratory analysis results were filled out after the clinical evaluation, and when available.
All samples were collected at Clinica Luganese Moncucco during hospitalisation and ICU stay.
All hospital records were registered in an electronic CRF. The CRF was filled in any time treatment or clinical outcomes changed. Any further record of the enrolled patients, such as results from radiological, cardiological, or pneumological exams, or others performed during the hospitalisation, was collected.
Follow-up
Participants were followed during hospitalisation and for 12 months after discharge. Samples were collected at hospitalisation, on Day 7, Day 14, and Day 21, at discharge, and at 1, 2, 3, 6, and 12 months after discharge. Additional samples were collected in case of worsening or amelioration of the clinical condition.
If the patient was transferred to a rehabilitation ward, the medical staff was contacted in order to collect the samples at the scheduled follow-up points.
Follow-up visits were scheduled at the outpatient Clinic of Epatocentro Ticino. If a patient failed to attend the appointment, he/she were sent at least two reminders via mail. If the patients did not attend an appointment after two further invitations, he/she were withdrawn from the project and listed as lost to follow-up.
LABORATORY PROCEDURES
The study physician was responsible for collecting clinical and laboratory data, and the study nurses were responsible for taking blood specimens. The collaborating laboratory, Medisyn Ticino, Switzerland, performed regular quality controls to ensure the accuracy of the results. At scheduled time points, filled blood test tubes were stored for the COVID-19 Biobank. During hospitalisation, blood samples were collected at Clinica Luganese Moncucco and, after discharge, at the Epatocentro Ticino. The specimens were handled and stored at Medisyn Ticino, a centralised, authorised laboratory for the handling of biosamples at biosafety level 2. The specimens collected and stored included sodium citrate blood, ethylenediaminetetraacetic acid blood, mRNA (PAXgene [PreAnalytiX GmbH, Zurich, Switzerland]) samples, serum, nasopharyngeal swabs, urine, sputum, stool, and tracheal aspirates.
Results were collected and entered in RedCAP for storage and further exportation and consultation.
Approval Process for Nested Projects
In order to gain access to clinical samples and data, researchers had to submit their research proposal to the scientific committee of the COVID-19 Biobank, which is composed of a panel of national experts in infectious diseases, virology, and immunology.
RESULTS
Patient Characteristics
Overall, 162 patients were eligible for the COVID-19 Biobank; 27 were excluded from enrolment for different causes, listed in the flowchart (Figure 1). In total, 135 patients with COVID-19 were enrolled, and a total of 10,116 samples were collected. The first patient was enrolled on 25th April 2020, and the last patient was enrolled on 13th December 2021. All patients signed the informed consent at the moment of hospitalisation.
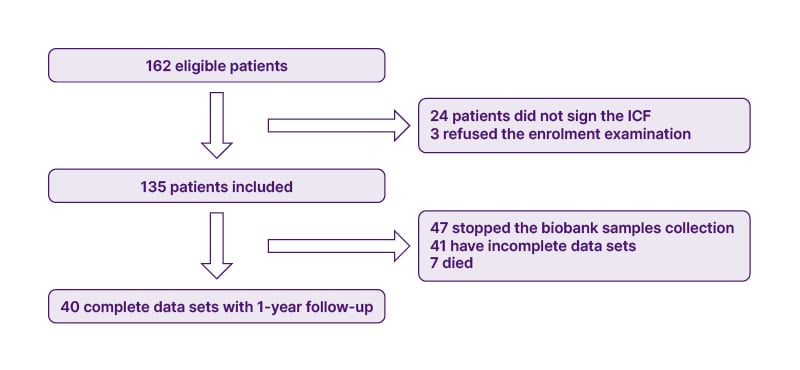
Figure 1: Study flowchart.
ICF: Informed consent form.
The infection was diagnosed with a nasopharyngeal swab at admission. As reported in Figure 2, 135 patients were hospitalised at the internal medicine ward, and two went directly to the ICU. Among those patients at the internal medicine ward, 22 had a clinical change. Of these, 16 patients underwent worsening of the clinical condition, which led 12 of them to be transferred to the ICU, of which one died. Another three patients died at the internal medicine ward (Figure 2). All the patients hospitalised in the ICU survived.
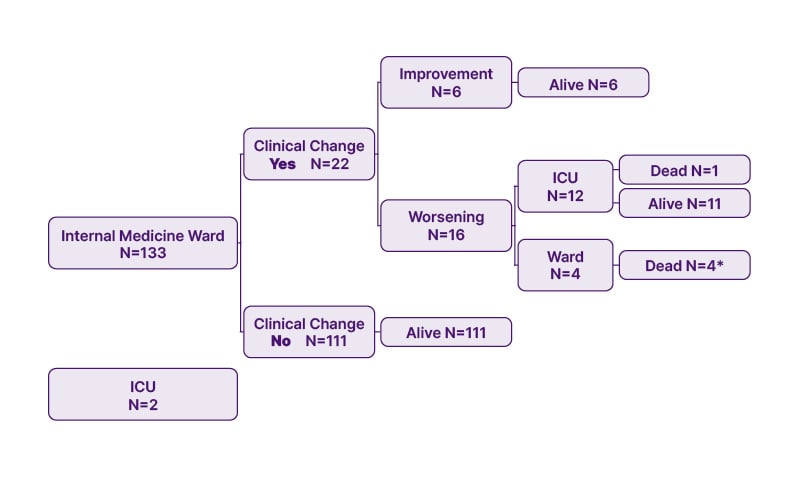
Figure 2: Patients’ characteristics.
*Two patients died after discharge.
Main characteristics are presented in Table 1. Briefly, most participants in the COVID-19 Biobank were male (68.9%) and White (98.5%). The mean age of patients was 65 years, and of these, 9.6% had previously had a SARS-CoV-2 infection.
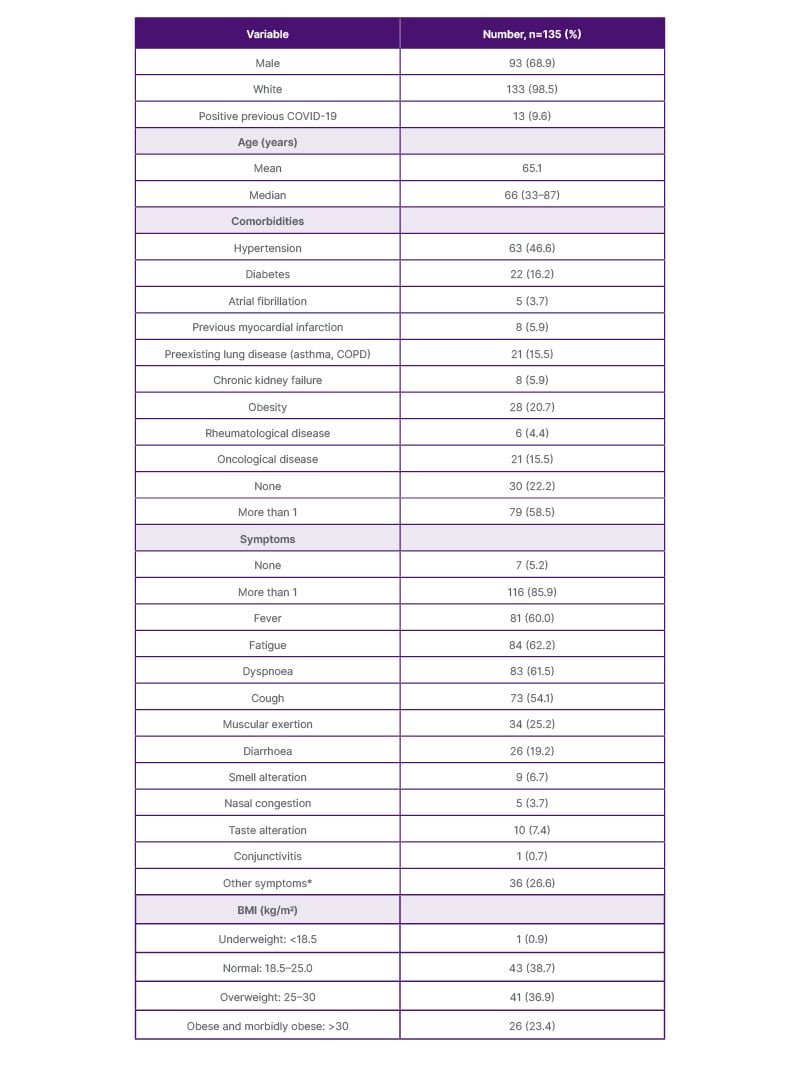
Table 1: Enrolled patients’ characteristics.
*Nausea, vomiting, abdominal pain, headache.
The authors registered the BMI value of 111 patients; 67 of them (60.3%) had a BMI >25 kg/m2.
Most patients presented with more than one symptom at hospitalisation. At onset, only seven (5.2%) patients displayed no symptoms. The most frequently described symptoms were fever (60.0%), fatigue (62.2%), and dyspnoea (61.5%).
During hospitalisation, 69 (51.1%) patients had a chest radiograph and 75 (55.5%) had a CT chest. The findings on radiography were focal thickening (50.7%) or bilateral interstitial infiltrates (31.9%). On CT scan, 52.3% of patients had pneumonia and focal thickening, and 22.7% had bilateral pneumonia (Table 2).
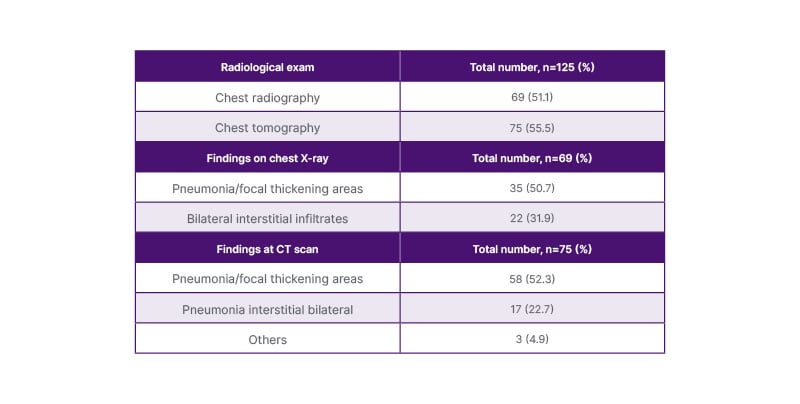
Table 2: Clinical and radiological records.
All patients provided samples at enrolment. During the follow-up period, the rate of adherence dropped to an overall 40%. The main decrease in adherence was registered during outpatient clinic follow-up. In total, there were 135 enrolment visits and 249 follow-up visits.
Forty-seven patients were asked to stop the sample collection, and 41 were lost to follow-up. The average age of the lost to follow-up group was 69 years (median: 73 years); 71% of them had more than one comorbidity. The main cause of drop-out or missing samples was a lack of interest in the study once discharged from the acute clinic, and the high number of samples required.
The cause of death in five out of seven patients was respiratory failure associated with SARS-CoV-2 infection; the other two patients died after discharge.
Laboratory Specimens
Figure 3 describes the specimen characteristics. The largest amount of retained samples were ethylenediaminetetraacetic acid and serum aliquots (15% of whole samples each), followed by nasal and oral swabs (14%), urine samples (12%), stool samples (8%), saliva samples (4%), and RNA samples (1%). The most requested samples for nested projects were the serum, RNA, and stool samples.
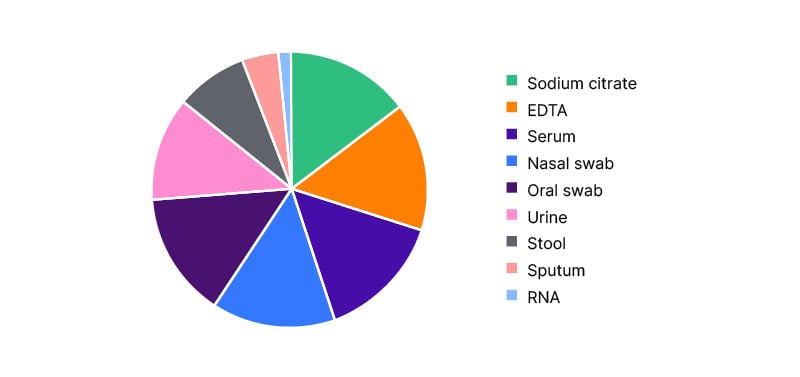
Figure 3: Specimen characteristics (%, N=3,679).
EDTA: ethylenediaminetetraacetic acid.
Nested Projects
This Biobank project was created with the primary objective of collecting data and samples from patients hospitalised with COVID-19 during the first wave of the SARS-CoV-2 pandemic. The aim was to obtain well-characterised, professionally collected, and stored samples, deposited in the COVID-19 Biobank alongside detailed clinical information of every single patient with their clinical evolution in order to allow investigators to carry out genetic, serological, microbiological, and immunological studies as nested projects.
All projects described had been approved by the scientific committee of the COVID-19 Biobank.
The first approved nested project is by Albrich et al.5 This project comprises a multicentre study aimed at understanding the interaction between gut microbiota and the host immune and metabolic systems that influence COVID-19 outcomes. The researchers performed a multi-omics analysis on the Biobank samples and compared those who had a fatal outcome (n=41) to those with severe but non-fatal outcomes (n=89), or mild/moderate diseases with non-fatal outcomes (n=42).
They identified eight cytokines and 140 metabolites in the sera of patients who had a fatal outcome, together with elevated levels of multiple pathobionts and lower levels of protective or anti-inflammatory microbes. They also found metabolism patterns that were correlated with a fatal outcome, together with the presence of pathobionts such as Enterococcus spp. In patients with less severe outcomes, they identified clusters of anti-inflammatory microbes Bifidobacterium spp or Ruminococcus spp, short fatty chain acids, and IL-17A. The second approved nested project was by Hensen et al.6 The aim of this project was to characterise the serum metabolomic trajectories of 71 patients hospitalised because of moderate or severe COVID-19. The study included three Swiss hospitals. Researchers found statistically significant differences in the serum metabolite concentrations of 444 out of 901 metabolites studied. The findings included markers of hospitalisation and markers of physiological functioning that might play a role in worsening lung injuries.
The third nested project is by Petkidis et al.7 The primary objective of this study was to develop an AI framework for ready detection of virus-induced cytopathic effect. They used the convolutional neural network EfficientNet-B0 (Google AI, Mountain View, California, USA), and transmitted light microscopy images of infected cell cultures, including SARS-CoV-2. They found out that the cytopathic effect induced by each virus is highly specific and provides unbiased infectivity scores of infectious agents.
Another study conducted with the COVID-19 Biobank samples was published in 2021.8 The authors formed a multinational network of researchers spread across the world to investigate the role of human genetics in COVID-19 symptoms and disease severity. They presented the results of a meta-analysis that included more than 49,000 patients from 46 studies across 19 countries. They found 13 genome-wide significant loci that are associated with a severe infection. Some of them correspond to already known associations between the genome and autoimmune and inflammatory diseases.
DISCUSSION
The COVID-19 Biobank is the first collection of clinical and epidemiological data on patients with COVID-19 hospitalised in Southern Switzerland. Its primary aim was to collect high-quality, well-annotated biological samples and clinical data. This objective has been achieved through rigorous standardisation of data collection and centralised storage of samples at Medisyn Ticino under the supervision of Fondazione Epatocentro Ticino. By ensuring consistent sampling protocols across hospitals and the Epatocentro Ticino and implementing double-entry validation of data, the Biobank has generated a dataset that is reliable, comprehensive, and suitable for both immediate and future research applications.
Importantly, this cohort covers a critical period of the pandemic, from the first wave (first patient enrolled 25th April 2020), when over 90% of patients were SARS-CoV-2 naïve, through the emergence of clinically significant viral variants (last patient enrolled 13th December 2021). This has allowed the collection of a unique set of naïve samples, providing an invaluable resource for studying disease outcomes, viral biology, and immune responses without the confounding effects of prior infection or vaccination.
In the broader Swiss and European context, the COVID-19 Biobank complements other initiatives such as the Swiss Biobanking Platform, providing a region-specific, longitudinal resource with intensive sampling during acute hospitalisation. By integrating clinical, epidemiological, and microbiological data, the Biobank strengthens collaborative opportunities and contributes to a more comprehensive understanding of COVID-19 at both national and international levels.
The study faced several limitations inherent to pandemic conditions. Frequent sampling of hospitalised patients was challenging due to high patient volumes, rapid clinical changes, and the strain on hospital personnel, which occasionally led to missed samples. Follow-up adherence declined over time, with only 40 of 135 patients completing the 1-year visit. Patient comorbidities and the milder disease course during later pandemic waves further reduced follow-up compliance. Despite these challenges, the Biobank retains a robust dataset and substantial sample repository suitable for ongoing and future studies.
FUTURE DIRECTIONS
Beyond COVID-19, the Biobank provides a platform for investigating other respiratory infections, immune responses in naïve versus previously exposed individuals, and long-term sequelae such as long COVID-19. Its standardised collection and centralised storage facilitate multicentre collaborations and nested studies, making it a valuable resource for both translational and epidemiological research.