BACKGROUND
Profiling of DNA methylation, a stable epigenetic marker of cell identity, has become a significant tool in precision cancer diagnostics, since DNA methylation reflects both somatically acquired changes and the cell-of-origin.1 Conventional diagnostics of brain tumours integrate histology, immunohistochemistry (IHC), and molecular classification with Infinium MethylationEPIC array (EPIC; Illumina, San Diego, California, USA) and gene panel sequencing.
However, these techniques require sample multiplexing for cost efficiency, leading to 2–4 weeks delay in obtaining integrative diagnosis, postponing tumour board and precision therapy initiation.2
METHODS
To shorten this time span, the authors analysed 45 native brain tumour samples using real-time nanopore sequencing. The goal was to implement a robust nanopore wet-lab workflow for clinical practice (Figure 1A), and evaluate accuracy of molecular classification in comparison with integrated neuropathological diagnosis.3 DNA was isolated, quantified, and sequenced on a MinION (Oxford Nanopore Technologies, UK) device. Data analysis was performed using the NanoDx™4 pipeline developed by Philipp Euskirchen (Charité Berlin, Institute of Neuropathology, Germany) and the Sturgeon5 pipeline developed by C. Vermeulen, M. Pagès-Gallego, L. Kester et al. (Oncode Institute, Utrecht, the Netherlands).
RESULTS
The authors found a tumour class prediction in 31 cases (71%) using the NanoDx classifier, and in 39 cases (89%) using the Sturgeon classifier. For NanoDx, the predicted molecular class was concordant with integrated neuropathological diagnosis in 29 cases (94% of classifiable cases), and in 29 cases for Sturgeon (74% of classifiable cases; Figure 1B).
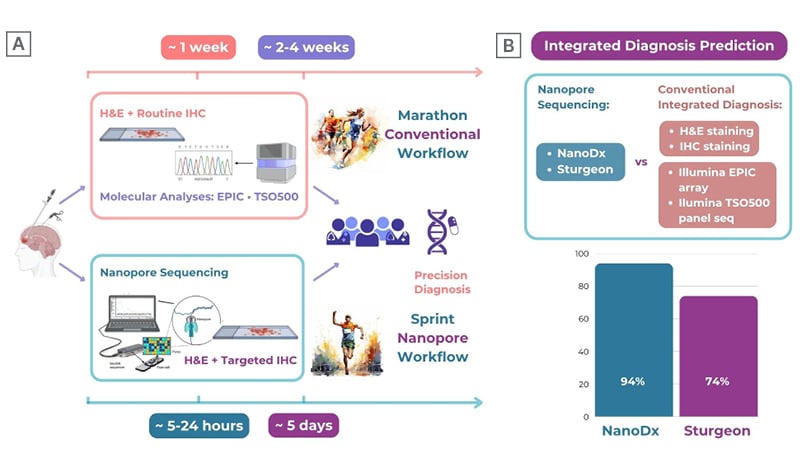
Figure 1: Diagnostic workflow and evaluation of the accuracy of molecular brain tumour classification with
nanopore sequencing at the Division of Neuropathology and Neurochemistry, Medical University of Vienna, Austria.
A) Conventional workflow (top) and novel nanopore sequencing-based diagnostic workflow (bottom) for neuropathological analysis of brain tumours.
B) Summary of the accuracy of tumour type prediction using nanopore sequencing (NanoDx and Sturgeon classifiers)compared to conventional integrated diagnosis.
Parts of the figure were generated using BioRender.
EPIC: Infinium MethylationEPIC array (Illumina, San Diego, California, USA); H&E: haematoxylin and eosin; IHC: immunohistochemistry; vs: versus.
CONCLUSION
The authors’ analysis demonstrates that nanopore workflow shortens molecular classification of brain tumours, reducing diagnostic timing from 2–4 weeks to approximately 24 hours.
This rapid profiling enables targeted IHC, facilitating an integrated tumour diagnosis potentially within 5 days, dramatically improving the timeline for precision therapy onset. Ongoing research aims to improve reliability by continuing to collect data and refine classifiers, implement targeted nanopore sequencing of specific genomic regions, like MGMT promoter, and assess the impact of faster integrated diagnosis on clinical outcomes.