Abstract
Asthma is characterised by episodic bronchospasm, airway hyperreactivity, and airway inflammation. Current treatment is aimed at reversing bronchospasm with bronchodilators and decreasing airway inflammation with corticosteroids. Asthma patients as a collective group, however, have variable responses to treatment, and our understanding and view of asthma as a single pathologic process has evolved substantially. We now recognise that asthma is a heterogeneous disease with many phenotypes, as reflected by differences in natural history, complexity, severity, and responses to treatment. The underlying aetiologies for many phenotypes are poorly understood and likely multifactorial. Recent evidence increasingly supports an important role for microbial exposures and our microbiota as factors mediating asthma pathogenesis. However, given the phenotypic heterogeneity of asthma, we further propose that microbiota may play an additional role in shaping asthma phenotype. Beginning with a brief overview of concepts of asthma phenotypes and endotypes, the intent of this article is to summarise current knowledge of the microbiome in asthma, highlighting recent studies that have examined relationships between microbiota and phenotypic features of asthma. We conclude with a discussion of future research directions, considering important issues and challenges in this area of investigation.
FROM PHENOTYPE TO ENDOTYPE: ASTHMA IS NOT ONE SIZE FITS ALL
Multiple asthma phenotypes have been described1,2 using frameworks that incorporate demographic, clinical, and inflammatory features. To describe known biology and more precisely define phenotypes, the term ‘asthma endotype’ was proposed in a 2011 consensus report published by the European Academy of Allergy and Clinical Immunology (EAACI) and the American Academy of Allergy, Asthma and Immunology (AAAAI).3 Fundamentally, an ‘endotype’ is a subtype of a condition defined by a distinct functional or pathophysiologic mechanism.3 It also has been proposed that asthma endotypes should be distinct in several parameters, including clinical characteristics, biomarkers, lung physiology, genetics, histopathology, epidemiology, and treatment response.3
Recent molecular studies of airway gene expression patterns have helped define at least one asthma endotype based on markers of T helper Type 2 (Th2) inflammation. Two of these studies examined bronchial epithelial cell gene expression patterns in asthmatic subjects,4,5 identifying sets of interleukin (IL)-13-inducible epithelial genes (POSTN, CLCA1, and SERPINB2) that characterised subjects who responded favourably to inhaled corticosteroids. A similar approach was adopted in a recent study of sputum cell expression of Type 2 and non-Type 2-related genes.6 Sputum-based gene expression profiling also molecularly identified subgroups of ‘Type 2 high’ versus ‘Type 2 low’ asthma, in particular expression of IL-4, IL-5, and IL-13. These studies, in conjunction with earlier data on the high prevalence of asthmatic subgroups with non-eosinophilic inflammation, have helped establish Type 2 high asthma as an endotype that characterises only ~50% of patients.7,8 A component of the definition of this endotype is the greater likelihood of responsiveness to corticosteroid therapy.5
However, not all Type 2 high asthma is responsive to steroid therapy alone. For instance, some patients with severe asthma have ongoing poor asthma control and airway eosinophilia despite high doses of inhaled corticosteroids.9 It is now known that Type 2 cytokines derive from other cellular sources besides classical Type 2 lymphocytes (e.g. innate lymphoid cells),9,10 and thus not all Type 2 immune responses may be steroid-sensitive. In this setting, new immunotherapeutics targeting specific cytokines that drive Type 2 inflammation (e.g. anti-IL-5, anti-IL-13, and anti-IL-4Ra) will become important adjunctive therapies in patients with evidence of ongoing Type 2 or eosinophilic inflammation despite steroid therapy.11
In contrast, Type 2 low asthma likely encompasses several sub-types of asthma with different underlying mechanisms, though these remain poorly understood. Thus the term Type 2 low asthma in itself does not define a particular endotype, but a common feature is poorer responses to corticosteroid therapy. Understanding mechanisms of Type 2 low asthma has been identified as a research priority.12 Although markers of Type 2 inflammation run a continuum, classifying asthma as Type 2 high versus Type 2 low is useful both clinically and in research. While the best approach to identify such patients in clinical practice remains undetermined, the distinction facilitates 1) appropriate tailoring of available treatments that largely target Type 2 inflammation,8,11 and 2) focussing of research efforts on mechanisms and treatment targets for Type 2 low phenotypes of asthma.
Despite the goal of asthma endotyping schemes, overlap among endotypes in clinical and inflammatory features is certain. Broad characteristics like timing of disease onset, gender, the presence of atopy, and level of lung function are common across describedphenotypes with different underlying mechanisms.13-16 For example ‘late-onset asthma’ includes several phenotypes.17 Among these, aspirin-exacerbated respiratory disease represents one of the best studied endotypes,18 characterised by later onset of asthma, chronic rhinosinusitis with nasal polyps, and acute respiratory reactions following the ingestion of non-steroidal anti-inflammatory drugs. Although eosinophilic sinus disease is prominent, the pathogenesis of aspirin-exacerbated respiratory disease is thought to involve inflammatory lipid mediators and cyclo-oxygenase pathways of arachidonic acid metabolism,18 rather than Type 2 immune responses specifically.
Other phenotypes of late-onset asthma are little understood in comparison.1,17 A distinct subgroup of adult-onset asthma patients have recurrent severe exacerbations with pulmonary function often lower than in allergic asthmatic patients, and the presence of sputum eosinophilia despite atopy being less common.19,20 Obesity-associated asthma is also a significant problem. However, adult-onset asthma complicating obesity may result from a different interplay of mechanisms versus childhood asthma, complicated by obesity. Studies of adult and paediatric cohorts have implicated both non-Type 2 and Type 2 pathways in subsets of obese asthma patients.21-23 Additional phenotypes characterised by low or absent Type 2 inflammation include those with neutrophil-predominant (non-eosinophilic) airway inflammation that is not solely attributable to concurrent use of inhaled corticosteroids.7,24 Potential aetiologies include chronic infection by bacteria and smoking-induced airway inflammation. Moreover, among patients on corticosteroids, it is quite possible that this superimposed therapy further shapes both the inflammatory and microbial milieu of the airways,25-28 complicating the understanding of potential asthma endotypes at this end of the disease spectrum.
In summary, many asthma phenotypes and potential endotypes exist but the underlying mechanisms for many of these remain unclear, especially in Type 2 low disease. It is likely that multiple factors shape the development of asthma phenotypes, such that the ultimate definition of any one endotype will contain clinical and/or inflammatory criteria that overlap with other endotypes. In addition, links between patterns of microbiota composition and specific features of asthma have been reported.26-29 Table 1 presents a non-comprehensive list of recent literature on asthma phenotypes, including studies that have focussed on the lower respiratory microbiome in asthma. We believe that as knowledge advances on how microbial factors shape asthma, the microbiome will become an additional domain necessary to consider and incorporate into efforts to define asthma endotypes.
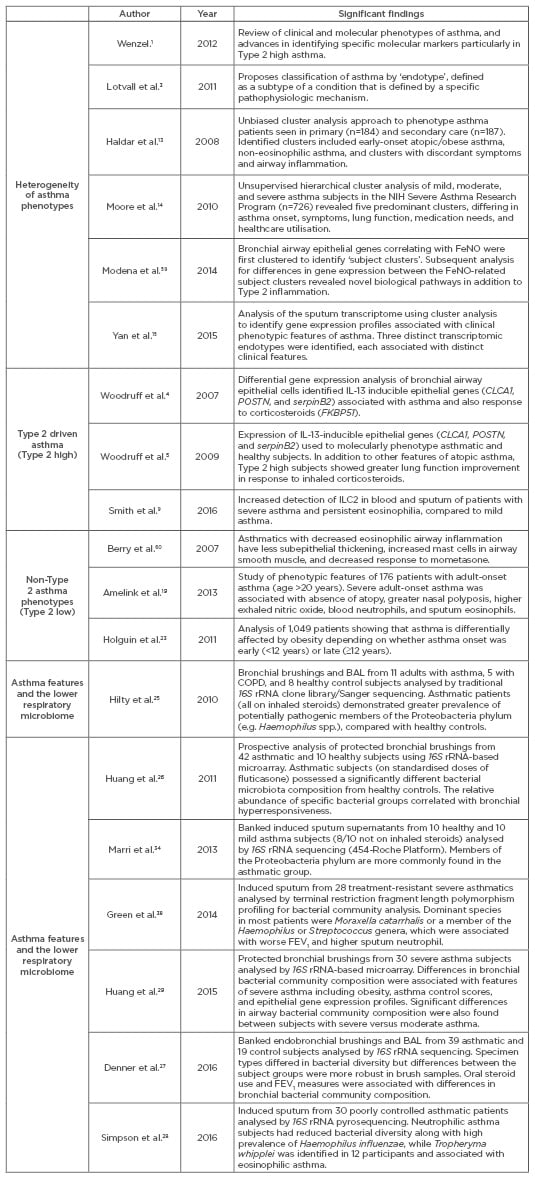
Table 1: Abbreviated list of publications highlighting the heterogeneity of asthma, Type 2 high versus Type 2 low phenotypes, and recent studies of the lower respiratory microbiome in asthma.
NIH: National Institutes of Health; COPD: chronic obstructive pulmonary disease; BAL: bronchoalveolar lavage; IL: interleukin; FeNO: fractional exhaled nitric oxide; ILC2: Type 2 cytokine-producing innate lymphoid cells; FEV1: forced expiratory volume in 1 second.
This parallels a recent editorial by Stappenbeck and Virgin30 who argue that we can no longer ignore the fact that “mammals are defined by their metagenome, a combination of host and microbiome genes,” and that this knowledge must be incorporated into efforts to understand human disease pathogenesis and disease phenotypes.
THE MICROBIOME AND ITS POSSIBLE ROLE IN ASTHMA PHENOTYPE
Study of the role of microbes in asthma has been of long-standing interest and is of increasing importance, as newer techniques to molecularly identify microbes advance knowledge in this area. Interactions between the microbial environment and the naïve immune system are key in the development of immune defences and determine the repertoire of the immune system.31 An emerging body of evidence shows that the microbial environment plays a role in the pathogenesis of atopic diseases, including asthma.32 Asthma has been associated with distinct differences in the composition of gut and respiratory microbiota, compared with healthy individuals.33-37 Moreover, different patterns of respiratory microbiota composition are associated with different phenotypic features of asthma,26-29,38 suggesting that microbial dysbiosis could play a role in asthma endotype(s). Specifically, features like airway hyperresponsiveness, airflow obstruction, obesity, and level of asthma control have been found to correlate with particular features of airway microbiota composition.26,27,29
Environmental Microbial Exposures and Asthma in Children
The association between a deficiency in environmental microbial exposure and increased prevalence of atopic disease is the basis of one of the earliest prevailing hypotheses on the pathogenesis of allergic disease. The ‘hygiene hypothesis’ is based on the principle that lack of early exposure to infectious agents or microbes that stimulate protective immune responses increase susceptibility to allergic diseases. Children raised in homes with multiple siblings39 or pets have decreased prevalence of atopic disease.40,41 Increased family size, exposure to day care, and increased crowding have also been associated with decreased rates of allergy or asthma.42-44 Asthma prevalence is greater among urban children, compared with their counterparts raised in rural areas and farming environments42-44 who may more likely be exposed to domesticated animals, outdoor microbes, and additional environmental exposures that shape immune responses. Though exact mechanisms remain incompletely understood, recent molecular insights have implicated endotoxin-induced modification of communication between airway epithelial and dendritic cells.45
Connections Between the Gut Microbiome and Asthma Development
It is estimated that 500–1,000 different bacterial species inhabit the mature gastrointestinal tract, with bacterial cells outnumbering host cells 10-fold.46 The micro-organisms colonising the gut perform different functions vital for human health, including processing of dietary constituents, regulation of host metabolism,47 and immune system maturation, including development of immune tolerance.31 Differences in birth mode and antibiotic exposures have been linked to alterations in gut microbial ecology and immune development.46 Studies utilising high-throughput molecular platforms have reported strong associations between gut microbiome features, immune responses, and the tendency to develop allergy or asthma. A comprehensive overview of these studies is beyond the scope of this article, and we refer readers to other reviews on this topic.33
In a study by Abrahamsson et al.,35 47 infants had stool samples collected at 1 week, 1 month, and 12 months of age. Seven years later, the children were assessed for allergic disease and skin-prick test reactivity. Lower microbiota diversity was found at 1 week and 1 month in children who went on to develop asthma by 7 years of age. A more recent study by Arrieta et al.36 similarly found that differences in gut microbiota composition in very early life were associated with greater risk of having atopic wheeze later in childhood. Decreased relative abundances of Lachnospira, Veillonella, Rothia, and Faecalibacterium in the first 100 days of life were seen among infants at high-risk of developing asthma in childhood. Another study showed that infants colonised at 3 weeks of age with Bacteroides fragilis group and/or Clostridium coccoides subcluster XIVa, had increased risk of asthma at 3 years of age.37
In addition to their immunostimulatory effects, gut bacteria also express crucial enzymes that permit metabolism of otherwise indigestible polysaccharides and dietary starches, leading to the production of short-chain fatty acids (SCFA), such as butyrate, propionate, and acetate that can be used for host ATP synthesis or synthesis of other energy substrates in the liver.47 SCFA are known to be important to colonic epithelial and mucosal health.47,48 Recent studies also suggest a role for gut-derived SCFA in modulating allergic airway inflammation.49
In contrast to paediatric studies or animal models of asthma pathogenesis, the potential role of the gut microbiome in adults with asthma has not been directly studied. Obesity is an important comorbidity of asthma and has been associated with increased asthma severity that is less steroid-responsive. Independent of asthma, the distal gut microbiota in obese humans or mice appears to be altered compared with lean states.50,51 Specifically, obesity has been associated with less diverse gut bacterial communities, with reported alterations in the ratio of Firmicutes to Bacteroidetes, the two most common bacterial phyla present in the gut.51 Feeding mice a diet high in fermentable fibre content altered the ratio of these two bacterial phyla, increased levels of circulating SCFA, and through their alteration of dendritic cell capacity to promote Type 2 cell effector function, protected against allergic inflammation in the lung.49 These in vivo studies provide a framework for understanding possible links between the gut microbiome and allergic inflammation in the lung.
THE AIRWAY MICROBIOME AND ASTHMA
The properties of the lower airway microbiome are distinct. The bacterial make-up of the lower airway is different from the upper airway and does not completely reflect oropharyngeal flora.52,53 While there is variability in the composition of lower airway microbiota from person to person, several studies have shown that the lower airway microbiome differs in the setting of chronic airway disease, compared with healthy state. For example, Proteobacteria, a large phylum that represents many known potential respiratory pathogens, are relatively more abundant in subjects with asthma and other obstructive airway diseases, compared with healthy controls.54 Members of the Proteobacteria more prevalent in airways of asthmatic subjects include the genera Haemophilus, Moraxella, Neisseria, and members of the Enterobacteriaceae family.25-29,34,38 Differences in respiratory microbiota composition have also been observed as early as the first year of life, mostly with nasopharyngeal sample analysis. In a prospective cohort of 234 children in Australia,55 nasopharyngeal samples collected from infants at age 2 months, 6 months, and 12 months were analysed by 16S rRNA gene sequencing to evaluate nasal bacterial microbiota composition. During this time, all incidents of acute respiratory infection in the children were documented. The infant nasopharyngeal microbiomes were dominated by six genera: Moraxella (31.2%), Streptococcus (15.5%), Corynebacterium (13.5%), Staphylococcus (10.3%), Haemophilus (9.7%), and Alloiococcus (8.8%). During acute respiratory viral infections, known to be associated with increased propensity to develop asthma,56 Moraxella, Streptococcus, and Haemophilus were more prevalent. In adjusted multivariate analyses, early asymptomatic colonisation with Streptococcus was found to be a strong predictor for current wheeze at 5 years of age as well as subsequent chronic wheeze at 10 years of age. These findings suggest that important viral-bacterial-host interactions occur that likely impact immune responses leading to asthma.
Microbiome diversity has been found to differ in paediatric asthmatic patients compared with healthy controls. In a single-centre observational study, nasal epithelial cells were collected from eight children with asthma and six healthy controls ranging ages of 6–20 years. RNA sequencing was used to pursue microbial identification.57 The nasal microbiome of asthmatic patients was less diverse when compared with healthy controls. Furthermore, there were higher levels of Moraxella in asthmatics. Escherichia and Psychrobacter (a member of the Moraxellaceae family) were found in greater abundance in nasal epithelial cells of asthmatic children compared with controls. Haemophilus influenzae, Streptococcus spp., and Staphylococcus spp. were detected in samples from asthmatic patients, although the relative abundance was not statistically significantly different compared with healthy controls.
Differences in the airway microbiome have also been observed across asthma of different severities.29 Phenotypic characteristics such as body mass index, measures of airway hyperresponsiveness, and asthma control have also been found to correlate with different patterns of lower airway bacterial microbiota composition.26,27,29 Moreover, a subset of asthma patients may harbour particular bacterial microbiota and be more likely to respond to treatment with macrolide antibiotic therapy. In a post hoc analysis of moderate asthma subjects randomised to treatment with clarithromycin for 16 weeks, those demonstrating an improvement in airway hyperresponsiveness after macrolide treatment had higher bacterial diversity at baseline.26 Studies have taken a closer look at which organisms are associated with greater asthma severity.24,25 A study of sputum bacterial composition in 28 treatment (steroid)-resistant asthmatic patients found that the relative abundance of Moraxella catarrhalis, Haemophilus spp., or Streptococcus spp. correlated with worse lung function, higher sputum neutrophil counts, and IL-8 concentrations.38 In a study that analysed protected bronchial airway brushings,29 similar patterns of increased proteobacteria were seen with greater asthma severity, with a subset of severe asthma patients demonstrating significantly greater relative abundance of Klebsiella (pneumoniae or oxytoca species). Concurrent steroid treatment may influence these findings, and further studies are needed. Taken together, however, existing evidence suggests that patterns of microbiome diversity or the relative abundance of particular groups of organisms, may serve as a microbial marker of patients more likely to respond to antimicrobial or other microbiome-targeted therapies in the future.
CONCLUSION AND FUTURE DIRECTIONS
Recent studies have elucidated the existence and complexity of the upper and lower respiratory microbiomes, and their associations with asthma including phenotypic clinical and inflammatory features. These extend earlier studies that focussed on the role of environmental microbial exposures or gut microbiota in asthma development. There are unique challenges to studying the lower respiratory microbiome, however. Direct sampling requires invasive bronchoscopy that is burdensome, costly, poses some risks, and hampers abilities to conduct large clinical studies in the outpatient setting. Recent studies have clarified that the upper and lower respiratory tracts harbour distinct compositions of bacterial microbiota,52,53 a difference magnified in the setting of chronic obstructive airway diseases.54 Nonetheless, studies of larger numbers of patients are important because significant heterogeneity between individuals in microbiota composition has been observed, regardless of health or disease status and even among those with advanced lung disease due to cystic fibrosis or chronic obstructive pulmonary disease.54,58 Longitudinal studies are also needed to understand the intrinsic stability or variability of the respiratory microbiome, both compositionally and functionally. We view this as a necessary foundation to link microbiome relationships to clinical outcomes and events (e.g. exacerbations) and identify potential microbial markers of disease endotype. Therefore, microbiome studies of other specimen types will be useful to gauge their utility, and potentially even their advantages over bronchoscopy-collected samples, to understand how respiratory microbiota influence asthma. There is precedence as quality-controlled sputum has been used for decades to understand inflammatory patterns in asthma, and recent analyses of nasopharyngeal samples have generated new hypotheses about microbiota relationships to asthma in children.55
We believe the following topics also merit further attention: 1) determining relationships between respiratory microbiota and patterns of host inflammation, which may help to define particular endotypes; 2) exploring whether alterations in the gut microbiome are associated with asthma in those with established disease; and 3) understanding what functions are elaborated by respiratory microbiota to shape host responses in asthma. Moreover, studies of the respiratory microbiome to date have largely focussed on bacteria due to well-established methods and the accessibility of techniques to study mixed populations of bacteria. Fungal and viral communities also are of interest, but molecular methods to analyse the mycobiome and virome, respectively, are more challenging than for bacteria. Other -omic approaches such as metagenomics (shotgun DNA sequencing), metatranscriptomics, and metabolomics can be leveraged to investigate the functional potential of members of the microbiota. However, applying these methods to study human respiratory samples, often of limited material volume, presents some challenges. Important choices may need to be made upfront regarding sample handling and processing for such analyses.
Asthma is a complex and heterogeneous disease whose pathogenesis, including that of different phenotypes, likely involves multiple components and pathways. The microbiome in itself is also complex, and its relationships with the host may be viewed as a network of interactions. More research is clearly needed, but emerging evidence suggests that microbial dysbiosis of the respiratory tract has the potential to shape manifestations of asthma and may underline particular disease endotypes.





