Abstract
The discovery of a new generation of highly potent broadly neutralising antibodies (bnAb) has provided a new weapon in the fight against HIV-1. It is envisioned that multiple bnAb or a single bnAb in conjunction with antiretrovirals (ARV) can be used to treat HIV infection, especially individuals harbouring extensively drug-resistant virus or those that require regimen simplification. Furthermore, it is believed that bnAb may eliminate latently infected cells through antibody-mediated cellular cytotoxicity, and this functionality may induce virus remission. BnAb epitopes and HIV envelope determinants for CCR5 and CXCR4 usage often overlap, and this provides the basis for believing that there is a relationship between receptor utilisation and bnAb sensitivity. This review highlights the important intersection between HIV co-receptor usage and bnAb therapy. Compared to CCR5-using strains, CXCR4 strains are generally more resistant to bnAb that target the V1-V2 apex and V3 N332 glycan, but not the other envelope domains. This association between bnAb sensitivity and co-receptor usage can be leveraged both to develop pre-treatment assays to identify resistant strains, as well as to anticipate potential adverse outcomes with future HIV antibody-based therapeutics.
INTRODUCTION
Combination antiretroviral therapy (cART) suppresses HIV-1 replication below detectable levels in the plasma, delays the onset of AIDS, and prolongs the lifespan of infected individuals.1 Current therapies, however, must be taken on a daily basis and continued indefinitely. Whenever antiretrovirals (ARV) are stopped, the virus begins replicating again, and the march toward AIDS resumes for the patient. Virus replication re-emerges because of the presence of infectious virus in latently infected cells which are not eliminated by cART.2 Chronic cART use can also be complicated by drug-associated adverse events and emergence of drug resistance. Thus, novel therapies are needed to overcome these shortcomings of current cART.
Passive infusions of some broadly neutralising antibodies (bnAb) or vaccination with vectors that generate antibodies have been shown to decrease virus replication in animal models3-7 and human trials.8-13 Antibody-based therapies provide an attractive new anti-HIV-1 weapon because of dosing simplification and activity against multidrug-resistant HIV. Furthermore, it has been hypothesised that antibody-associated effector functions, such as natural killer cell-mediated antibody-dependent cellular cytotoxicity, can eliminate latently infected cells, which could potentially induce virus remission in the absence of ARV.14,15 Often, the same HIV envelope glycoprotein domain contains both a bnAb epitope and the determinant for receptor usage that is necessary for host cell entry. This overlap provides the scientific basis for speculating that there is an association between the receptors viruses use to enter cells and bnAb sensitivity. Although there have been a number of recent reviews on bnAb, there has been relatively sparse discussion of the intersection between HIV-1 receptor utilisation and bnAb sensitivity.16-19 The primary purpose of this review is to discuss the association between receptor usage and neutralising antibodies (nAb) and to highlight possible clinical implications for future antibody-based treatments.
BROADLY NEUTRALISING ANTIBODIES TARGETING ENVELOPE VARIABLE LOOP DOMAINS MAY IMPACT RECEPTOR USAGE
Developing strategies that obviate the need for daily ARV, either through a sterilising cure that eliminates all latently infected cells or a functional cure that prevents virus re-emergence after cART discontinuation, are top priorities in the HIV-1 field.2 The isolation of potent and broad nAb from HIV-1-infected individuals has energised this endeavor.16-19 Even though no immunogen has been designed that can elicit bnAb, repeated passive bnAb infusions or vaccination with a vector that produces an antibody are being proposed as feasible alternative strategies to generate therapeutic levels that suppress plasma viraemia and induce antibody-mediated cellular cytotoxicity that can potentially eliminate latently infected cells. In contrast to the current ARV, bnAb can be given monthly or at even longer intervals. Furthermore, bnAb have been shown to enhance the anti-HIV-1 humoral immune response in animal models; this augmented immunity could yield virus remission in absence of ARV.14
In order to enter cells, HIV must bind the CD4 receptor and then a co-receptor: either CCR5 or CXCR4.20 Early after infection the majority of HIV-1-infected individuals harbour variants that bind the CCR5 receptor (termed R5). Over time, some individuals can develop a dual-mixed (DM) virus population containing a mixture of R5 variants, viruses that are capable of using both the CCR5 and the CXCR4 (R5X4), or exclusively the CXCR4 (X4) receptor.21-23 The evolution to CXCR4 usage over the course of HIV infection primarily depends on sequence evolution in the HIV envelope glycoprotein variable loop 3 (V3). Envelope glycoprotein V3 loop modifications are often accompanied by modifications in the variable loop 1 and 2 (V1-V2) region due to fitness constraints.24,25 Although the HIV envelope glycoprotein sequence changes over time in all infected individuals, not everyone develops CXCR4-using virus. In addition, the percentage of individuals that eventually develop a DM population varies by HIV-1 subtype.21-23 Among the most prevalent HIV-1 subtypes circulating in the world, people infected with HIV-1 subtype B (HIV-1B) and D (HIV-1D) are more likely to have CXCR4-using virus as compared to those infected with subtypes A (HIV-1A) and C (HIV-1C). In addition to having differences in the frequency of CXCR4-usage, various subtypes also have varying susceptibilities to different bnAb directed against the envelope.26 Envelope glycoprotein differences among the various HIV-1 subtypes contribute to the variation in both bnAb sensitivity and frequency of CXCR4-using variants. The overlap between the location of the antibody epitopes and the co-receptor usage determinants potentially provides a rationale for these observations.
The majority of nAb against HIV-1 attach to the HIV envelope and prevent host cell entry by restricting receptor attachment and subsequent fusion. Currently the isolated bnAb target diverse envelope regions, including 1) the CD4 binding site (bs); 2) glycosylation-dependent epitopes in the V1-V2 loops; 3) a glycan patch encompassing the envelope V3 loop; 4) membrane proximal external region on the envelope gp41 domain; 5) glycan-dependent epitopes that bridge gp120 and gp41 including the fusion peptide; and 6) the silent face.27-34 The V1, V2, and V3-directed bnAb target a glycosylated asparagine (N) at envelope position 160 and 332 respectively, and thus they are often also referred to as N332 and N160 bnAb, respectively. While the activity of the N160 and N332 bnAb primarily depend on the presence of these glycans, other amino acids, especially those in and around the V1-V2 and V3 envelope regions, also impact neutralisation. For instance, the N332 bnAb PGT121 interacts with residues at the tip of the V3 loop, including the crown and amino acids directly before and after it. Another N332 bnAb, 10-1074, interacts with residues towards the base of the V3 loop.35 Involvement of these other envelope V1-V2 and V3 amino acids, similar to those identified as key determinants for co-receptor usage, provides the basis for speculating that co-receptor usage may be associated with bnAb activity.
When considering this association, it is crucial to understand the differential neutralisation sensitivity of CCR5 and CXCR4-using viruses. Earlier studies implied that CXCR4-using viruses were more antibody neutralisation-sensitive compared to CCR5-utilising strains. In an animal model, it was demonstrated that the emergence of CXCR4-using simian HIV was temporally related to the loss of humoral immunity.36 Antibodies that were present prior to the emergence of the CXCR4-using virus potently neutralised the X4 strain. This suggested that antibodies had prevented the emergence of a CXCR4-using virus and the X4 strain appeared as an escape variant only after humoral responses diminished with progressive immunodeficiency. The earliest human studies also deemed CXCR4-utilising strains more neutralisation-sensitive compared to CCR5-utilising strains. These investigations primarily compared the neutralisation susceptibility of either co-circulating or heterologous CCR5 and CXCR4-using strains to heterologous antibodies, such as first-generation bnAb, which have relatively limited potency and breadth compared to second-generation bnAb.37-40 More recent studies have provided some conflicting data. First, the administration of second-generation N332 bnAb (PGT128) after established infection in humanised mice generated resistant virus.6 Importantly, envelope sequence but not phenotypic analysis predicted that a higher proportion of the emerging resistant virus used the CXCR4 as opposed to the CCR5 receptor.41 This implies neutralisation-resistant CXCR4-using variants emerged with N332 bnAb, PGT128, administration. In contrast to earlier human studies which never assessed sensitivity to second-generation bnAb, X4 as compared to R5 strains are less neutralisation-sensitive to N160 (PG9 and PG16) and N332-directed bnAb (PGT128 and CAP8 serum).41,42 It should be noted that these studies have not evaluated the possibility that a greater frequency of X4 as compared to R5 isolates lack the N160 or N332 glycan.
This is unlikely, however, because the absence of N160 and N332 have never been demonstrated to be predictive of CXCR4 usage. Furthermore, recent structural data demonstrates that the envelope 332 amino acid makes no contact with the CCR5 receptor (Figure 1).43 The structure also suggests that the envelope 160 amino acid does not directly interact with the CCR5 receptor although the solved structure lacked this V1-V2 amino acid. It has been suggested that X4 as compared to R5 strains have a more open trimer structure, and this change may prevent N160 bnAb from binding because these antibodies depend on a quarternary epitope.28-44 The X4 and co-circulating R5 variants, however, have similar susceptibility to CD4 bs (VRC01 and VRCPG04) and membrane proximal external region (10E8)-directed antibodies. Furthermore, co-circulating X4 as compared to R5 strains have been shown as more neutralisation-resistant to contemporaneous autologous and heterologous plasma.42 In aggregate, the earliest and newer studies suggest that X4 and R5 variants have different neutralisation sensitivities. Importantly, the most recent studies have shown that X4 compared to R5 strains are more resistant to the second-generation bnAb that target the N160 and N332 envelope glycans.
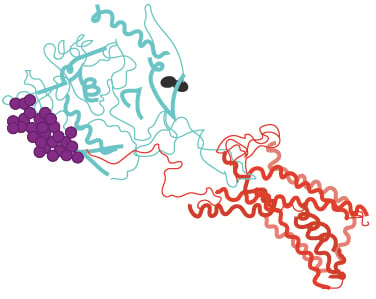
Figure 1: Structural model of HIV-1 envelope and CCR5 receptor.
Structural model shows HIV-1 envelope (cyan) and CCR5 receptor (red). Amino acid 332 is highlighted in black. The V1-V2 loop is shown is purple. The original structure lacked the envelope 160 amino acid. The published structure (PDB 6meo) was manipulated in PyMol for this figure.43
HUMORAL IMMUNE PRESSURE MAY SELECT FOR CXCR4-USING VIRUS
The mechanism for the evolution of DM virus population and the difference in the frequency for the emergence of CXCR4-using variants in various subtypes remains uncertain. Although the distinction between CCR5 and CXCR4-utilising strains has been known for over 25 years, surprisingly, previous studies have not elucidated a definitive mechanism for the emergence of CXCR4-using viruses over the course of infection. One hypothesis suggests that CXCR4-using viruses potentially emerge because of inadequate CCR5 receptor levels or limited availability of CCR5 bearing target cells.22 Indeed, individuals with heterozygous CCR5 delta 32 genotype, who have lower levels of the CCR5 receptor on their cells, have an increased likelihood of harbouring CXCR4-using viruses compared to people with both wild-type alleles.45 It should be noted, however, that after CXCR4-using strains emerge, R5 strains often persist, implying that co-receptor switching does not always occur in response to the limited availability of CCR5 receptor positive-susceptible cells.21,23 As stated above, the observations from the non-human primate studies argue another possible hypothesis; CXCR4-using variants arise because of random mutations, and these new viruses persist because declining adaptive and innate immune responses fail to clear the less fit dual and/or X4 HIV-1.22,36
Lastly, newer observations suggest yet another possibility, that humoral immune pressure may play a role in the emergence of CXCR4-using viruses. While it is true that all HIV-1-infected individuals develop nAb against their autologous circulating viruses, the envelope region targeted by the autologous antibodies differs based on the characteristics of the circulating strains.46-50 Indeed, only a small proportion of HIV-1-infected individuals develop a broad and potent humoral immune response, and the evolution of a bnAb is directly dependent on the antigenic properties of the circulating viruses’ envelope glycoprotein.51-54 The authors speculate that some infected individuals likely develop a unique type of antibody, termed CXCR4-inducing antibody (CXCR4-iAb) (Figure 2). The emergence of a CXCR4-iAb only occurs in the presence of specific HIV envelope glycoproteins and predominate only in certain HIV-1 subtypes. Because glycosylation modifications in the envelope variable loops have been associated with the R5–X4 transition, it is possible that CXCR4-iAb target glycan-rich epitopes in the envelope V1-V2 or V3 regions.21,24,25 To escape these nAb pressures, some R5 variants could evolve CXCR4 usage while other co-existing R5 viruses that are not susceptible to CXCR-iAbs retain the same co-receptor phenotype. This speculative model predicts that individuals with CXCR4-iAb should have X4 variants that are more neutralisation-resistant compared to co-existing R5 variants. As stated above, previous studies have already demonstrated multiple instances in which co-circulating CXCR4 compared to CCR5-using variants are more neutralisation-resistant to autologous plasma.42 The model further predicts that passage of some neutralisation-sensitive R5 variant in the presence of CXCR4-iAb should lead to the emergence of the CXCR4-using virus.
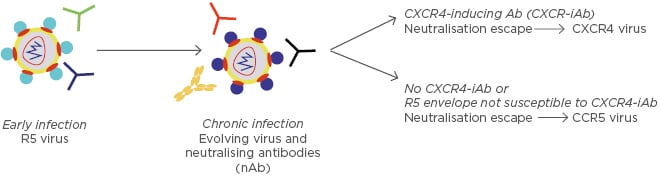
Figure 2: Potential model for emergence of CXCR4-using strains.
CCR5-using viruses (primarily R5) are present early after virus acquisition. Over time, the virus envelope glycoprotein changes in response to neutralising antibodies. CXCR4-using viruses emerge in some individuals that develop unique antibodies termed CXCR4-iAb. Individuals that lack CXCR4-iAb continue to have CCR5-using strains.
Indeed, this was observed with the emergence of a CXCR4-utilising variant after passaging a R5 variant in the presence of autologous contemporaneous plasma in vitro.42 Furthermore, neutralisation-resistant CXCR4-using strains have been shown to emerge late in disease, suggesting that these types of strains emerge only after prolonged exposure to host humoral immune pressure.55 Isolation and characterisation of these proposed CXCR4-iAb will provide definitive support for this proposed mechanistic model. It should be noted, however, that there is no direct evidence from human passive infusion clinical studies that a N160 or a N332 glycan bnAb induces co-receptor switching.
There is a body of other data also supporting the notion that co-receptor switching may occur as a consequence of neutralisation escape. For instance, HIV-1B, HIV-1C, and HIV-1D X4 compared to co-circulating R5 variants often have distinctive V3 loop motifs.21,23 The X4 strains in these subtypes often have a 2–3 amino acid V3 loop insertion in the same general V3 loop region (Figure 3). Yet, the forces promoting V3 insertions remain unclear. The similarity in the V3 loop insertions among HIV-1B, HIV-C, and HIV-1D X4 variants suggests that these highly divergent viruses are independently converging to a similar solution to escape a common selection pressure, likely nAb. Indeed, nAb selective pressure has been associated with insertions observed in V1 through V4 envelope domains.46-48,56 This data argues that humoral immune pressure selects for X4 variants with the observed V3 loop insertions because there is low likelihood that random mutations will lead to a shared genotypic and phenotypic characteristic. Thus, neutralisation escape is hypothesised as one mechanism among the diverse non-mutually exclusive postulated processes that potentially explains co-receptor switching.22
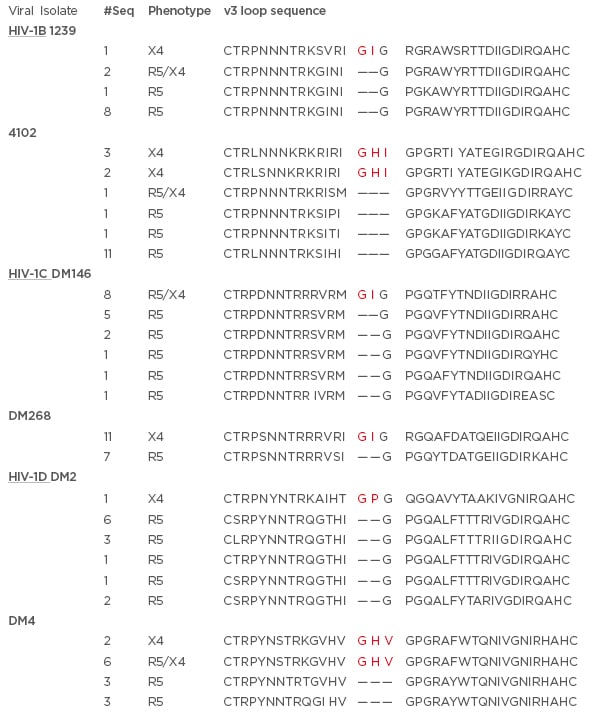
Figure 3: Representative examples of co-circulating HIV-1B, HIV-1C, and HIV-1D R5, R5X4, and X4 strains.
The alignment of the predicted amino acid sequence (denoted by single letter abbreviation) of phenotypically confirmed X4, R5, and R5/X4 envelope variable 3 (V3) loops. The subtype is listed and underlined on the left. Columns indicate the subject ID, number of envelopes with the same predicted V3 loop sequence, and the confirmed co-receptor phenotype. Bold red letters denote the predicted insertions. HIV-1C and HIV-1D sequences are from previous publications21,23 while HIV-1B sequences are from a manuscript under consideration.
CLINICAL IMPLICATIONS FOR THE INTERSECTION FOR THE INTERSECTION BETWEEN BROADLY NEUTRALISING ANTIBODIES THERAPY AND CO-RECEPTOR USAGE
The intersection between HIV co-receptor usage and the future use of bnAb as HIV therapy has a number of important clinical implications. First, the association between co-receptor usage and differential bnAb sensitivity to N160 and N332 loop directed-bnAb could be leveraged for determining the presence of neutralisation-insensitive viruses prior to treatment. Prior to implementing cART, infected individuals are examined for the presence of drug-resistant variants using sequence-based assays.57 Similar pre-treatment tests are not available to test susceptibility for these future antibody-based therapies. Extensive sequence variation in the HIV-1 envelope has made it difficult to develop a genotype-based test for predicting decreased antibody neutralisation sensitivity.
Individuals that contain variants with envelopes that lack the predicted glycan at position 160 or 332 are resistant to N160 and N332-directed bnAb, respectively. There are numerous HIV strains, however, that have decreased sensitivity to the N160 and N332 loop-directed bnAb even though they contain the required predicted glycosylated amino acids.58 In the absence of reliable sequence-based tests, bnAb clinical trials have often employed phenotypic assays. This entails the laborious process of examining neutralisation sensitivity of virus stocks incorporating HIV envelopes isolated from patient samples.10,12 Patient viruses are generated either through outgrowth cultures or in vitro virus stock preparations using envelope amplification, cloning, and transfection. Furthermore, these phenotypic methods lack sensitivity because outgrowth cultures and in vitro virus generation mostly capture major variants but not minor yet clinically important strains present in the patients’ viral quasispecies.59 On the other hand, numerous sequence-based methods can accurately predict co-receptor usage.60-62 It is possible that an infected individual’s circulating virus envelope sequence can be interrogated with high resolution using next-generation sequencing, and subsequently the receptor usage can be predicted through previously defined sequence-based algorithms. The predicted presence of CXCR4-using virus would imply decreased sensitivity of the virus population to N160 and N332 bnAb, based on the association between co-receptor usage and bnAb susceptibility as highlighted above. This sequence-based test would limit the use of bnAb among patients that harbour relatively neutralisation-insensitive viruses, which would prevent both the virus from developing resistance to other drugs in a combination cocktail and unnecessary costs.
The second important clinical implication of the link between co-receptor usage and antibodies is the theoretical possibility that N160 and N332 bnAb-based therapies may promote the persistence or emergence of CXCR4-using strains. This outcome would be clinically significant because presence and emergence of CXCR4-utilising viruses is associated with worse and more rapid disease progression.63,64 The potential for favouring the emergence of CXCR4-using virus with the use of N160 and N332-directed bnAb needs to be seriously considered prior to its widespread use because the therapeutic use of these bnAb may paradoxically worsen rather than improve disease outcomes. In humans and animals treated with N332-directed bnAb (10-1074 and PGT121), variants often escape neutralisation pressure by eliminating the glycan at 332 amino acid position, but this outcome is not universal.3,4,7,10-12 It has been suggested that changes to the 332 glycan may yield a strain that is highly susceptible to other autologous antibodies present in chronically infected individuals.65 Thus, in the presence of N332-directed bnAb, viruses may explore other sequence paths towards resistance, such as evolving toward CXCR4-usage. As opposed to in vitro passage, more pathogenic, phenotypically confirmed R5X4 or X4 strains have not been observed to emerge with N332 bnAb treatment. However, it is possible that CXCR4-using viruses evolve only after multiple rather than a small number of antibody infusions, which would be more consistent with clinical observations that these viruses emerge later in disease. Because of the association of X4 viruses with poorer prognosis and more rapid clinical progression, avoiding either the selection for a pre-existing or the emergence of X4 variant with bnAb treatment is clinically crucial. Viruses resistant to traditional ARV often have decreased replication capacity, and thus their emergence is not associated with worse disease outcome. In contrast to these ARV resistant strains, emergence of CXCR4-using viruses in the presence of specific bnAb would potentially lead to greater morbidity because co-receptor switching has been associated with faster disease progression.63,64 Furthermore, emergence of CXCR4-using strains would eliminate the ability to treat patients with CCR5 receptor inhibitors. Indeed, the CCR5 inhibitors are considered an alternative second-line agent in cART regimens, and thus the loss of this ARV class will also limit options for ARV salvage regimens. The potential for selection or emergence of CXCR4-using virus with use of bnAb will require further examination as bnAb enter the clinical sphere.
CONCLUSIONS
N160 and N332-directed bnAb are in clinical trials, and these novel therapeutics will likely enter clinical practice in the near future. The overlap of the HIV envelope domain targeted by these antibodies with the envelope regions involved in co-receptor usage provides the scientific rationale for believing that HIV variants that use different co-receptors to enter cells likely have varying susceptibility to these bnAb. Indeed, newer studies show that X4 compared to R5 HIV are less sensitive to N160 and N332 bnAb, and neutralisation pressure, such as during bnAb-based treatment, may favour the persistence and emergence of CXCR4-using strains. The authors speculate that the linkage between co-receptor usage and susceptibility to variable loop-directed bnAb can be exploited to develop assays to predict pre-treatment neutralisation sensitivity, as well as give insight into the potential complications with future N160 and N332 bnAb.






