Abstract
Introduction: Carbapenem resistance in Gram-negative bacilli (GNB) is a major concern in the management of resistant infections. The mechanism of carbapenem resistance is most commonly mediated by carbapenemases. The five most common genes (NDM, KPC, VIM, OXA, and IMP) are responsible for carbapenemase production. Knowledge of these genes is important for the management of the disease.
Objective: To estimate the prevalence of different genes responsible for carbapenemase production in GNB at a tertiary healthcare centre in South India.
Method: In this retrospective study, samples were collected over 16 months. Carbapenem-resistant GNB underwent to Xpert Carba-R assay (Cepheid, Sunnyvale, California, USA) for the detection of five important genes responsible for carbapenemase production: NDM, KPC, VIM, OXA, and IMP.
Results: Out of 184 carbapenem-resistant GNB, 20 samples were not included in this study. The rest of the 164 samples grew Klebsiella pneumoniae (152), Escherichia coli (10), and Enterobacter (2). OXA-48 and NDM were the most common genes responsible, with 137 (84.5%) and 95 (58.6%), respectively. Among them, 70 (43.2%) showed the presence of both genes, and 1 (0.6%) showed the presence of OXA-48, NDM, and VIM. Individually, 66 (40.7%) of OXA-48, 24 (14.8%) of NDM, and one (0.6%) of VIM. In this study, the authors did not find the presence of IMP or KPC genes.
Conclusion: As a result of limited options and the higher cost of antibiotics for carbapenem-resistant infections, knowledge of these genes helps in the selection and rational use of antibiotics reduces the cost of management and will prevent mortality and morbidity from these infections.
Key Points
1. To deal with multi-drug resistant bacteria, it is important to know the underlining mechanism of drug resistance. Molecular investigation is a new advancement in medical science and is useful in identifying this mechanism at a molecular level.
2. Carbapenemase production is the most important mechanism responsible for carbapenem-resistant Enterobacteriaceae. Knowing the prevalence of genes responsible for this carbapenemase production is very important for the selection of antibiotics.
3. Knowledge of the prevalence of genes responsible for carbapenemase in a particular region will help in antibiotic stewardship for the selection of empiric antibiotics when advance laboratory investigations are not available or while waiting for the reports.
INTRODUCTION
Carbapenem-resistant Enterobacteriaceae (CRE) is defined as Enterobacteriaceae that are resistant to the carbapenem group of antimicrobials. In the case where bacteria are intrinsically resistant to imipenem, the resistance to at least one carbapenem other than imipenem should be there to classify the organism as carbapenem-resistant.1
Resistance to carbapenems in Gram-negative bacteria, especially Enterobacteriaceae, is a major concerning emerging issue. There is a rising prevalence in the number of cases of CRE worldwide. In its publication ‘Antibiotic Resistance Threats in the United States’, the Centers for Disease Control and Prevention (CDC)2 refers to CRE as an urgent threat that is claiming more than 1,000 deaths annually in the USA. Similarly in Europe, there are increasing reports of CRE, causing disseminated and hospital-acquired infections. The highest prevalence of CRE is observed in Mediterranean and Balkan countries. The prevalence in Greece and Italy is reported to be around 60% and 40%, respectively.3,4 According to a study done in Asia, prevalence of CRE infection ranges from 0.6–0.9% of the total culture-positive infections.5 Although there are no uniform data available from India regarding CRE, published articles show the prevalence of carbapenem resistance among Enterobacteriaceae in India ranges from 18–31%. As most samples were collected from hospitalised patients in tertiary care centres, it may not represent the entire population.6-10
In order to act effectively, carbapenem molecules need to cross Gram-negative bacterial cell wall and reach intramembranous space with the help of the porin channels. There are three major mechanisms by which Enterobacteriaceae become resistant to carbapenem: enzyme production (carbapenemases), the formation of efflux pumps, and porin channel mutations. Of these, enzyme production is the main mechanism of resistance.11 The porin channels can act as a filter and prevent antibiotics from reaching the site of action. The efflux pump removes the antibiotic molecule from the intramembranous space.12 Unlike other mechanisms, carbapenemase-mediated resistance is usually transmitted by plasmids. They can easily be transferred from affected bacteria to unaffected ones. There lies the public health importance of carbapenemase-mediated resistance to carbapenems.12
Knowledge of different genes producing carbapenemase is important to be able to choose the appropriate antimicrobial.13 Xpert Carba-R assay (Cepheid, Sunnyvale, California, USA) is a rapid, real-time PCR assay that is useful for identifying genes encoding for the carbapenemase along with bacterial isolates from various clinical specimens, and can be useful for managing patients with CRE. Among the carbapenemase encoding genes, NDM, KPC, VIM, OXA, and IMP are recognised as the most important carbapenemases because of their wide prevalence.14
The current study was undertaken to understand the prevalence of different genes responsible for the production of carbapenemases in the authors’ setting.
Objectives
The objectives of this study were to estimate the prevalence of different genes responsible for carbapenemase production among Enterobacteriaceae in a tertiary healthcare setup in Bangalore, India, and to outline the treatment considerations for a patient with CRE infections where advance investigations are not possible.
METHODOLOGY
A retrospective chart review with convenient sampling completed between January 2020–April 2021. The study took place at a single centre, tertiary care, multi-specialty hospital in South India.
Inclusion criteria
Inclusion criteria includes all CRE being isolated during the study period and treating physicians requested for the molecular study (Xpert Carba-R) for the further management of the patient.
Exclusion criteria
Exclusion criteria includes carbapenem resistant Gram-negative bacteria other than Enterobacteriaceae and repeat samples from the same patient during the same hospitalisation period.
Data Collection
Data were extracted retrospectively from the Department of Microbiology between January 2020–April 2021, a period of 16 months. All CRE positive culture reports (from blood, urine, cerebrospinal fluid, swabs, and other body fluids) where the Xpert Carba-R assay was performed were included in the analysis. In the authors’ laboratory, antibiotic testing is done by VITEK® 2 (bioMérieux, Marcy-l’Étoile, France) compact. Enterobacteriaceae that show resistance to carbapenems (defined by meropenem minimum inhibitory concentration: >8 mcg/mL)15 were subjected to the Xpert Carba-R assay. The Xpert Carba-R assay is performed using the GeneXpert (Cepheid) platform. This is qualitative in vitro real-time PCR assay that detects five important genes producing carbapenemases, which include IMP, KPC, NDM, OXA-48, and VIM. This is an automated method wherein the samples from the culture plate are vortexed at high speed for 10 seconds in an elution reagent tube, which is later transferred into the specimen chamber of the Xpert Carba-R assay cartridge, as per the manufacturer’s instructions. The result is interpreted by the machine and the run time is 47 minutes. One of the limitations of the Xpert Carba-R assay is that it is only able to detect four out of 10 variant genes (blaOXA-48, blaOXA-162, blaOXA-163, and blaOXA-204) of OXA-48.16 This assay has high sensitivity and specificity (100% and 77%, respectively), with a positive predictive value and negative predictive value of 96% and 100%, respectively.17
RESULTS
A total of 50,144 patients were hospitalised during the study period. Among them, 9,424 (18.80%) had culture-proven infections. A total of 4,713 (50%) of the culture-proven infections were caused by Enterobacteriaceae. Among the total hospitalised patients, 2.55% had CRE infections. Among the total culture positive infections caused by Enterobacteriaceae, 27.18% were CRE.
Out of 1,281 CRE positive samples, 164 samples were analysed based on the inclusion and exclusion criteria. Out of these 164 samples, 64.0% were blood culture samples (105 out of 164); 12.2% (20 out of 164) were urine samples; 9.1% (15 out of 164) were tracheal aspiration; and purulent discharges constituted 6.1% (10 out of 164), while the rest of the 14 samples were collected either from cerebrospinal fluid, wound swab, sputum, tissue samples, or other body fluids.
Klebsiella pneumoniae was the main Enterobacteriaceae isolated, constituting 92.64% (152 out of 164) of the organisms, while 10 samples (6.09%) grew Escherichia coli, and two samples (1.21%) grew Enterobacter.
The authors could identify the gene responsible for carbapenemase production in 162 samples by the Xpert Carba-R assay. However, two samples tested negative for all the identifiable genes (five genes) by the Xpert Carba-R assay, which could mean there were other enzyme coding genes or non-carbapenemase mechanisms responsible for the resistance.11
Among the CRE isolates, 40.7% (66 out of 162) showed the OXA-48 gene alone, while 14.8% (24 out of 162) showed isolated NDM gene alone and 0.6% (1 out of 162) showed VIM gene as the inducer of resistance. A total of 43.2% (70 out of 162) of isolates showed the presence of a combination of both OXA-48 and NDM genes, and 0.6% (one) organism showed the presence of OXA 48, NDM, and VIM in combination. Overall, the OXA-48 gene was present in 84.5% (137 out of 162) isolates and NDM gene was the second common identified gene, having been found in 58.6% (95 out of 162) isolates. Notably, all the isolates were negative for IMP or KPC genes.
Table 1 shows the common resistance genes identified in each of the isolated organisms. The most common resistance gene carried by Klebsiella species, which was the authors’ most common isolate, was a combination of OXA-48 and NDM, followed by OXA-48 alone and NDM alone. The most common resistance gene identified in E. coli species was the NDM gene in isolation, unlike the Klebsiella species.
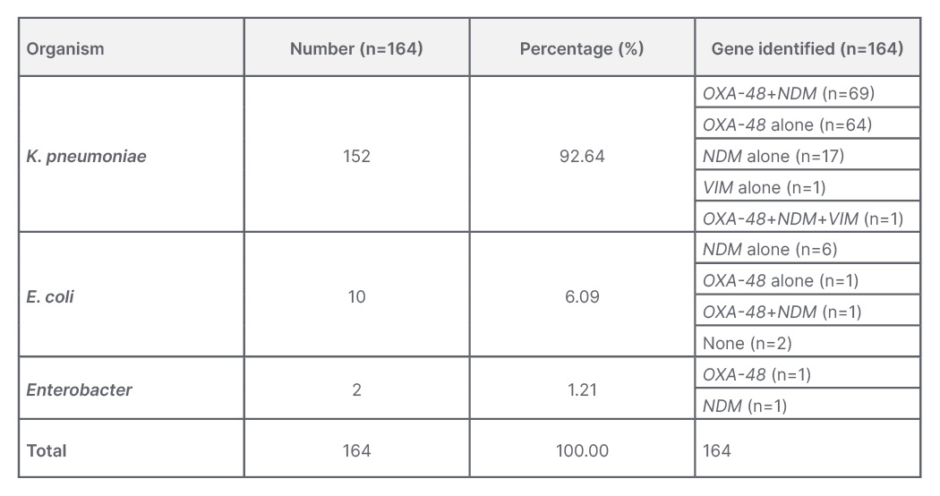
Table 1: Carbapenem-resistant Enterobacteriaceae organisms and identified genes responsible for carbapenem resistance.
E. coli: Escherichia coli; K. pneumoniae: Klebsiella pneumoniae.
DISCUSSION
Early, rapid identification of the genes causing carbapenem resistance will help to choose appropriate antibiotics early in treating CRE infections. In this study, 162 out of 164 samples could identify the genes for carbapenemase production. The remaining two samples could have had other genes (other than the five that are identifiable by the Xpert Carba-R assay) or could have carbapenem resistance due to non-carbapenemase mediated mechanisms.
Among CRE isolates, although the OXA-48 gene was the most prevalent gene in isolation (n=66 [40.7%]), which was followed by NDM (n=24 [14.8%]), the most common genetic mechanism causing carbapenem resistance was a combination of OXA–48 plus NDM, as noted in 43.2% (70 out of 162) isolates.
In the authors’ study, the most common CRE organism was K. pneumoniae (92.64% [152 out of 164]) followed by E. coli (6.09% [10 out of 164]]), and Enterobacter (1.21% [two out of 164]). Among the Klebsiella isolates, the most common mechanism of carbapenem resistance was found to be a combination of OXA-48 and NDM (45.40% [69 out of 152]), followed by OXA-48 alone (42.00%) and NDM alone (11.20%).
NDM alone (60% [6 out of 10]) was the most common mechanism recognised in E. coli, followed by OXA–48 alone (10%) and combination of OXA-48 plus NDM-1 (10%).
In two of the isolates of E. coli, no gene could be identified. With one–one samples being positive for both OXA-48 and NDM, both and were equally prevalent in the Enterobacter isolates.
In the study by Anandan et al.,18 the predominant organisms were K. pneumoniae (n=88) and E. coli (n=32) out of the 120 CRE isolates from blood cultures. Conventional PCR identified an equal number of isolates showing NDM genes (40.0% [n=48]) and OXA-48-like genes (39.2% [n=47]). E. coli was the predominant NDM producing gene (62.5% [n=30]), followed by K. pneumoniae (37.5% [n=18]). K. pneumoniae was the predominant isolate testing positive for OXA-48-like gene (83% [n=39]), followed by E. coli (17% [n=8]). However, a total of 15 (12.5%) carbapenem-resistant isolates were found to be coproducers of OXA-48 and NDM, unlike in the authors’ study, where the combination mechanism was found to be most predominant.18
In the study by Sekar et al.,19 89 isolates were E. coli out of 177 isolates of CRE, and 88 isolates were the Klebsiella species. Among the E. coli isolates that tested positive for the carbapenemase genes, the predominant gene was NDM (12 out of 33), followed by KPC (9 out of 33) and OXA-48 (6 out of 33). Among the Klebsiella species testing positive for the carbapenemase genes, 15 out of 32 were NDM, nine out of 32 were OXA-48, and three out of 32 were KPC, followed by two for VIM, two for NDM plus OXA-48, and one for NDM plus KPC.19 These observations again highlight the differences in the pattern of resistance mechanisms compared to the authors’ study.
Giri et al.20 completed a similar study, where they collected samples from a tertiary care hospital in western Maharashtra, a state in India, and found that 90% (n=45) detected the presence of NDM gene, 60% (n=30) showed the presence of OXA-48, and 12% (n=6) showed VIM gene in 50 isolated CRE samples. Their findings were similar to the results presented in this article by the authors, with the most common mechanism of carbapenem resistance found to be due to the combination of NDM and OXA–48 genes in 50% of the isolates, mostly associated with K. pneumoniae isolates.20
The study by Han et al.21 identified the KPC gene as the most common mechanism of resistance among their CRE Klebsiella isolates, followed by the NDM and OXA-48 genes; however, their E. coli isolates showed that NDM gene was predominant.
A multicentric study completed by Traczewski et al.22 across the USA and Europe found that, out of 467 isolates, Enterobacteriaceae (predominantly K. pneumoniae and E. coli) are the most common isolates (343 out of 467). followed by Pseudomonas aeruginosa (80 out of 467), and Acinetobacter baumannii (44 out of 467). This study also showed that among 343 samples of CRE, 89 of them showed the presence of OXA-48, 83 KPC, 73 NDM, 51 VIM, and four IMP-1.22
There are three molecular major classes of β-lactamases produced by Gram-negative bacilli: Ambler Class A, Class B, and Class D (Table 2). In this, there is one more chromosome-encoded cephalosporinases (Class C or AmpC), which is produced by these bacteria that may possess slightly extended activity towards carbapenems, but it is not clinically much significant.23
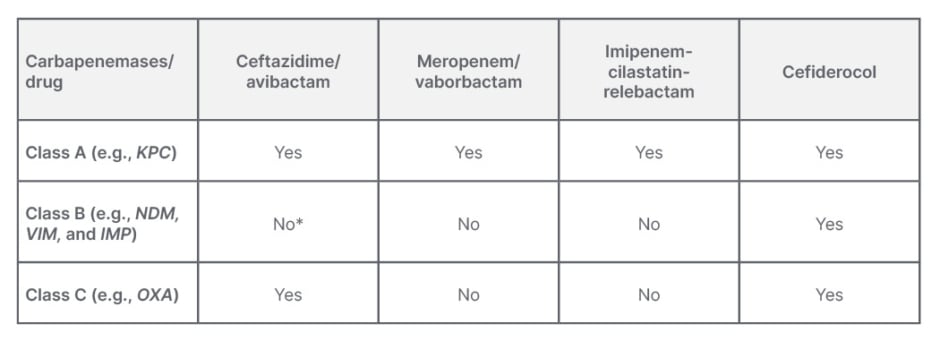
Table 2: Different carbapenemases and response to antibiotics.12,13,23,24
*Broad spectrum of hydrolytic activity including all penicillins, cephalosporines, and carbapenems will not work with exception of monobactam aztreonam.24,25
Three major types of Class A carbapenemases include the nonmetallocarbapenemase class A/imipenemase, serratia marcescens enzyme, and K. pneumoniae carbapenemase enzymes. This class of enzymes hydrolyses a broad variety of β-lactams, which includes penicillins, cephalosporins, carbapenems, and aztreonam. But newer β-lactamases are showing effectiveness against them.13,23
Class B is a metallo β-lactamases (MBL), which have a broad spectrum of hydrolytic activity against all penicillins, cephalosporins, and carbapenems. Commonly available β-lactamase inhibitors (such as clavulanic acid, tazobactam, or sulbactam) are also ineffective against this class of β-lactamases. But monobactam and aztreonam are showing activity against them. The NDM, VIM, and IMP genes encode these enzymes.13,23
Class D β-lactamases are oxacillinases (OXAs); they include more than 200 enzymes, among them a few variants possessing some carbapenemase activity. This class of β-lactamases are not able to hydrolyse expanded-spectrum cephalosporins.13,23
When looking at the CRE isolates from the index and other studies (Table 3) Klebsiella and E. coli are seen as the predominant organisms implicated in these infections. Also, what is more striking is the finding that the NDM gene is predominant, either in isolation or in combination with OXA-48, as the mechanism of resistance in most isolates irrespective of the organism. The understanding of the local antibiogram along with the likely organism suspected to be causing the nosocomial infection is very vital in choosing the appropriate empirical antimicrobial whenever healthcare professionals are faced with such nosocomial infections (Table 2). The current standard antimicrobial sensitivity tests done in laboratories can identify isolates with carbapenem resistance but are unable to identify the mechanism of carbapenem resistance. Understanding of the mechanism of carbapenem resistance is vital in deciding the appropriate antimicrobial (Table 2). However, there is a lack of utilisation of these molecular testing methods when faced with CRE infections due to various reasons, like a lack of availability or of clear understanding of their utility and needs.
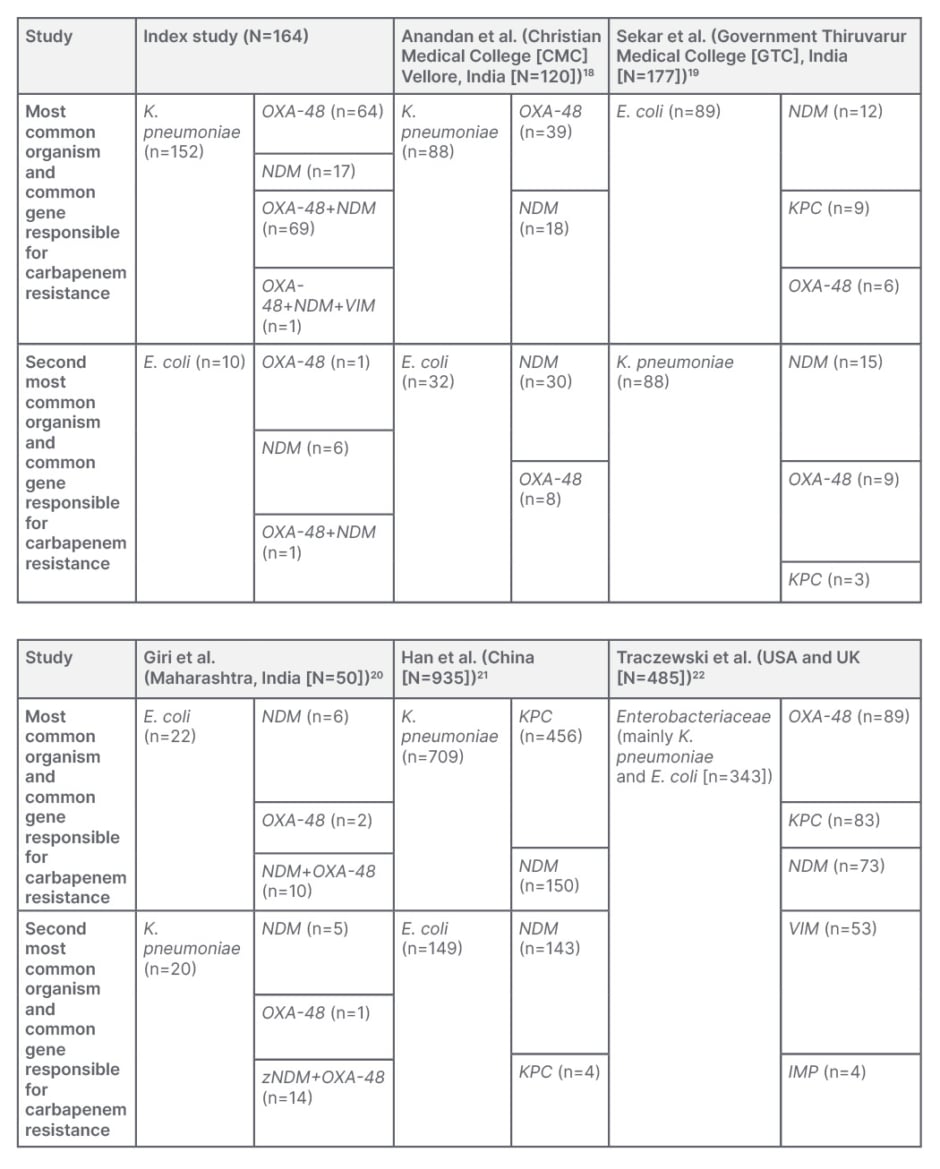
Table 3: A comparison between different studies.
E. coli: Escherichia coli; K. pneumoniae: Klebsiella pneumoniae.
The authors’ study could be used as some form of guidance in understanding the challenges and also in making the right choices when it comes to treating CRE infections. The NDM gene, being the predominant mechanism arming the CRE organisms, means that it becomes difficult to treat them adequately with the current available antimicrobial armamentarium. The ceftazidime avibactam combination is the only new antimicrobial combination currently available in the authors’ setting. This combination thankfully works for the OXA-48 strains. However, in the presence of the NDM mechanism of resistance, what is needed is the combination of aztreonam avibactam.24,25 In the absence of this combination in the market currently, the authors are forced to use the combination of ceftazidime avibactam plus aztreonam to tackle these nosocomial infections. What seems to be the way forward is for the authors to have the combination of aztreonam avibactam available, which will help them to spare the ceftazidime avibactam combination to treat OXA-48 infections alone. The concern is the emergence of resistance and, above all, what is more important is to realise the ever increasing means of resistant infections and working committedly towards various preventive measures that aim to limit such infections, and also the emergence of the resistance mechanism.
LIMITATIONS
The authors believe that the current study from their centre, being a single centre study, may not be representative of the prevalence of CRE infections and the mechanisms of resistance in these infections in every centre in this part of the globe. Hence, a multicentre study in a larger scale, across various parts of the country, is the need of the hour.
The current molecular testing employed in their lab (Xpert Carba-R) can identify limited mechanisms of carbapenem resistance. Two of their isolates testing negative for all known mechanisms suggest the possibility of other unidentifiable mechanisms that is outside the scope of the Xpert Caba-R assay. Availability of more advanced molecular testing, with a broader panel of genetic testing possibilities, will be able to identify those mechanisms, and may help the authors to understand and prepare them to tackle these organisms better.
Finally, this was a molecular study, but there were no Clinical and Laboratory Standards Institute (CLSI) guidelines available for the newer β-lactam/β-lactamase inhibitors combination antibiotic sensitivity pattern of the bacteria.
CONCLUSION
Appropriate molecular testing to identify the mechanism of resistance in CRE infections is important for understanding the epidemiology of these infections and plan the appropriate antimicrobial therapy.
Availability of cost-effective molecular testing facilities and improving awareness of the importance of testing are needed in order to tackle CRE infections more effectively.
KPC gene mechanism does not seem to be the prevalent mechanism of resistance among the authors’ CRE isolates at the time the study was undertaken. However, the authors may need to periodically look into the available data annually to recognise and change their antimicrobial prescription practice.
Moving forward, a properly planned multicentric study with adequate representative samples will be able to give a clearer understanding of the prevailing mechanisms of CRE resistance in India, and is the need of the hour.
Above all, every healthcare setup needs to have a written infection control policy and mechanisms in place to make sure there is the appropriate implementation of the same in order to prevent these life-threatening infections from affecting patient outcomes in the era where technological advancements have given us the opportunity to treat complex disease processes effectively.