BACKGROUND AND AIMS
Coexistence of hepatitis B virus (HBV) surface antigen (HBsAg) and antibodies (anti-HBs) can be found in 2.3–5.2% of chronic HBV (CHB) infected individuals. Recent reports have indicated that the presence of both biomarkers is associated with a high risk of hepatocellular carcinoma.1,2 However, previous studies with clinical samples presenting both biomarkers have demonstrated that the presence of variants in the HBsAg surface (S) gene and anti-HBs subtype can non-specifically contribute to the coexistence of these two biomarkers in CHB.3
METHODS
In this study, HBsAg variants were analysed in a clinical sample from a 43-year-old female with CHB, with prolonged (>7 years) coexistence of HBsAg and anti-HBs, HBV e-antigen-negative, and treatment with tenofovir disoproxil fumarate. Total DNA was purified from plasma and HBV DNA was amplified. Next-generation sequencing and a novel HBV-specific assembly/machine-learning tool for quasispecies were applied for the analysis of the S gene (GATACA Assembly Tool [GAT], Newport, Virginia, USA).
RESULTS
The authors determined the frequency of 17 site variants in the S gene (Table 1). Variants Y100S and L127P occurred together in the same haplotypes and eight of the sites studied presented with a variant with a stop codon. Additionally, several quasispecies of the same amino acid variation in the variants’ pool were found within this clinical sample. In addition, the study demonstrated that defective HBsAg variants can coexist with the wild type and other functional variants. Seven site variants were found in the ‘a’ determinant amino acid region of HBsAg, which could explain its persistence despite the presence of anti-HBs.
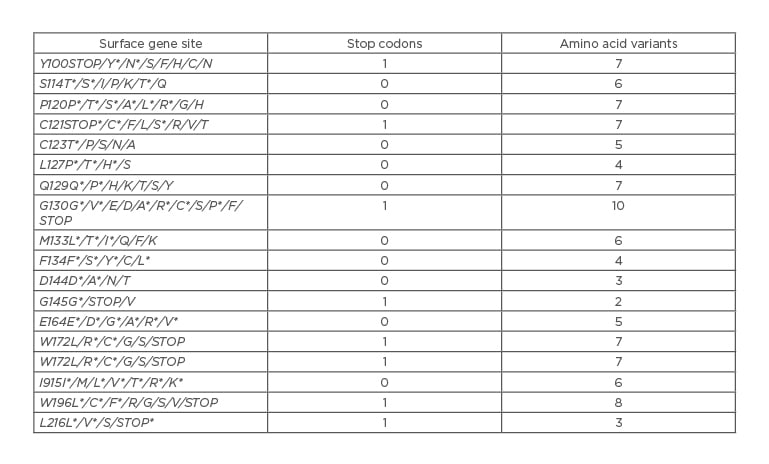
Table 1: Surface gene sites with amino acids identified with GATACA Assembly Tool (GATACA, Newport, Virginia, USA).
*Amino acid with different nucleotides.
CONCLUSION
Considering that the HBsAg S gene is a potential target for antiviral development and HBsAg seroconversion is a potential outcome for a functional cure, understanding the dynamics of these variants can help identify strategies of use for the novel direct-acting antiviral agents.

![EMJ Hepatology [Supplement 2] ILC Congress Review 2020](https://www.emjreviews.com/wp-content/uploads/2020/10/EMJ-Hepatology-8-Suppl-2-Feature-Image-940x572.jpg)