Abstract
Background: Pakistan has the second highest prevalence of hepatitis C in the world after Egypt. Viral hepatitis is a leading cause of morbidity and mortality in Pakistan and, worryingly, reinfection rates are also on the rise. This cross-sectional study was aimed at finding the most common genotypes of hepatitis C in terms of age and sex in a Pakistani cohort.
Materials and methods: The authors collected blood samples from 1,260 patients with diagnosed hepatitis C visiting a primary teaching hospital affiliated with Peshawar Medical College, Pakistan, from different districts of Khyber Pakhtunkhwa, Pakistan, between January 2017 and April 2019. Hepatitis C virus RNA was quantified by real-time polymerase chain reaction and genotyping was then performed.
Results: The authors found that genotype 3a was the most prevalent type followed by 1a, mixed, and 3b, respectively. Genotypes 2a and 1b were the least prevalent in Khyber Pakhtunkhwa. The most common genotype was 3a, observed in 75.87% of cases. The most common mixed genotype was 3a+1a, observed in 39 cases (3.10%); it had a prevalence of 3.49% in females compared with 2.70% in males. Overall, the most common age group affected by hepatitis C virus was 41–50 years (31.35%), followed by the 51–60 years group (24.45%). Infection rate was comparatively low in other age groups. A significant difference was observed in the prevalence of genotype 3a and 2a among different districts.
Conclusion: The authors concluded that genotype 3a was the most prevalent genotype and it was observed more frequently in the female population, with a median age of 45 years.
INTRODUCTION
The goal set by the World Health Organization (WHO) to eradicate hepatitis C by 2030 is a challenge for Pakistan, given that the prevalence of the virus is near that of an epidemic; a meta-analysis published in 2018 estimated a prevalence of 6.2%, which is the second highest worldwide after Egypt.1 Hepatitis C virus (HCV) is a single-stranded, enveloped RNA virus from the Flaviviridae family, genus Hepacivirus. The genome of HCV encodes a single polyprotein, which is processed co-translationally. The single polyprotein divides into three structural and seven non-structural polypeptides.2,3 These enclosed glycoproteins are responsible for the initial stages of infection in a host cell.4
At present, HCV has seven recognised genotypes and 67 subtypes. The genotypes are characterised by a discrete geographical dispersion and clinical expression; for example, genotype 1b is predominant in Japan, while in the USA and Europe genotype 1a is more common.5 The genotype is also correlated to age and transmission route; for example, individuals who use intravenous drugs are mostly reported to have HCV genotypes 3a and 1a, while genotype 1b is common in those who acquired the virus via blood transfusion.6 It was observed that the older population in the USA mostly acquired HCV iatrogenically, thereby showing a correlation with age. The number of individuals worldwide infected with HCV is 120–130 million, which is approximately 3% of the world population. According to the WHO, 3–4 million individuals in Pakistan are infected with hepatitis C each year, which contributes about 11 million HCV cases to the global disease burden.7,8
HCV infection is on a constant rise in Pakistan because of a lack of awareness, poor sterilisation techniques, reuse of contaminated and disposable syringes, and unscreened blood transfusions.9 Due to non-specific symptoms, it is difficult to pick early HCV infection, even in individuals with high viraemia. The positive cases identified in clinical surveys are usually in individuals with chronic hepatitis C.10 Acute HCV infection may lead to chronicity because of delay or irregularity in treatment. The majority of chronic cases of hepatitis C lead to cirrhosis and hepatocellular carcinoma.11 Due to poor socioeconomic circumstances, at present there is no health insurance system in Pakistan. The health department also receives a very small budget from the government and hence, the majority of patients still rely on cheap and easily available treatments, such as interferon and peginterferon.12 Until recently, direct-acting antiviral agents were extremely expensive; however, due to recent cost regulations, these therapies have become more widely available.13 Nevertheless, financial constraints still make it difficult for all of the 11 million individuals infected with HCV to be treated with direct-acting antiviral agents. Fortunately, the most prevalent genotype, 3a, responds effectively to the more conveniently available interferon therapy. Proper screening followed by genotyping is imperative before starting antiviral treatment as the efficacy, duration, and outcome of treatment will be dependent upon those tests.14,15
MATERIALS AND METHODS
Sample Collection
The authors conducted a cross-sectional study at Prime Teaching Hospital, Peshawar, Pakistan, affiliated with Peshawar Medical College, where patients from almost all districts of Khyber Pakhtunkhwa attend. A total of 1,260 HCV-positive cases, 630 males and 630 females, were included in the study between January 2017 and April 2019.
Virologic Examination
A total of 1,260 HCV-positive serum samples from an equal number of males and females (630 each) were selected for viral RNA extraction. INNO-LiPA (line probe assay) HCV kit (Innogenetics N.V. [now Fujirebio Europe N.V.], Ghent, Belgium) was used for the genotyping of HCV. Two sets of HCV universal biotinylated primers were selected for the amplification of the 5’ non-coding region of the HCV genome because of highly conserved base pairs. The HCV RNA-amplified products were hybridised to immobilise oligonucleotides specific for the genotypes of HCV. The HCV RNA quantification method described by Sarrazin et al.16 was used for CAP/CTM test (Roche Molecular Systems, Inc., Branchburg, New Jersey, USA). The extraction of RNA was performed by automated COBAS® AmpliPrep using 1 mL of plasma, and the extracted RNA was amplified by automated real-time polymerase chain reaction. The COBAS® TaqMan® 48 Analyzer was used for the detection of amplified RNA. Data analysis was performed using the AMPLILINK software, which presented HCV load as Log10 international units/mL. The real-time-PCR-positive samples were further subjected to genotyping assays.
Statistical Analysis
This study did not test any specific hypothesis. The continuous data were presented in percentages and descriptive statistics were outlined. The independent samples t-test and binomial test were used. A p value of <0.05 was considered significant.
RESULTS
Hepatitis C Virus Genotype Distributions in Different Age Groups
A total of 1,260 patients were divided into seven age groups. Starting from 10 years of age, each group had a difference of 10 years and HCV genotype prevalence was recorded and analysed for each group (Figure 1). The highest prevalence of HCV was observed in the 41–50 years age group with 395 cases (31.35%), followed by the 51–60 years age group with 308 cases (24.45%). The lowest HCV prevalence was noted in the patients >70 years of age (1.43%) and those <20 years of age (3.65%). There was no significant relationship found between age and genotype (p=0.028) when controlled for sex.
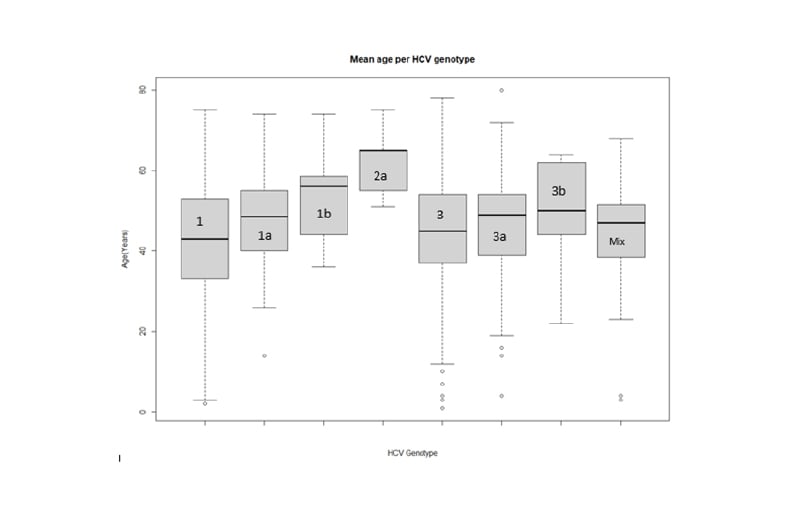
Figure 1: Box plot of distribution of mean age across hepatitis C virus genotypes. HCV: hepatitis C virus.
Sex-Based Hepatitis C Virus Genotype Distributions
It was observed that the most prevalent HCV genotype in both sexes was 3a, with a total of 956 cases (75.87%); however, the prevalence was slightly higher in females with 487 cases (77.30%) compared to 469 cases (74.44%) in the male population. The second most prevalent HCV genotype observed was 1a with 250 isolates (19.84%). Genotype 1a was, however, slightly more prevalent in males, with 135 isolates (21.43%) as compared to 115 isolates in females (18.25%). A fairly significant number (39; 3.10%) of isolates had mixed genotypes, with a slightly higher prevalence in females: 22 cases (3.49%) compared to 17 cases (2.70%) in the male population. Genotypes 1b, 2a, and 3b were more common in males.
The highest number of patients from a single district of Khyber Pakhtunkhwa was 736 (58.41%) from Peshawar, followed by Mardan with 78 cases (6.19%), Lower Dir with 63 cases (5.00%), and Dera Ismail Khan with 57 cases (4.52%). The most prevalent genotype in all districts was 3a, with slightly differing rates of prevalence. For example, district Buner had 94.12% 3a isolates, which was the highest prevalence of genotype 3a, followed by Nowshera with 91.12%, and Banu with 89.47%; Swabi had comparatively low prevalence of 3a with 57.89%. The second most common genotype, 1a, had the highest prevalence in Khyber Agency with 33.34% isolates, followed by Swabi with 31.58% and Mohmand Agency with 26.09% isolates. There were significant variations in the prevalence of HCV genotypes in the different districts of Khyber Pakhtunkhwa (Figure 2).
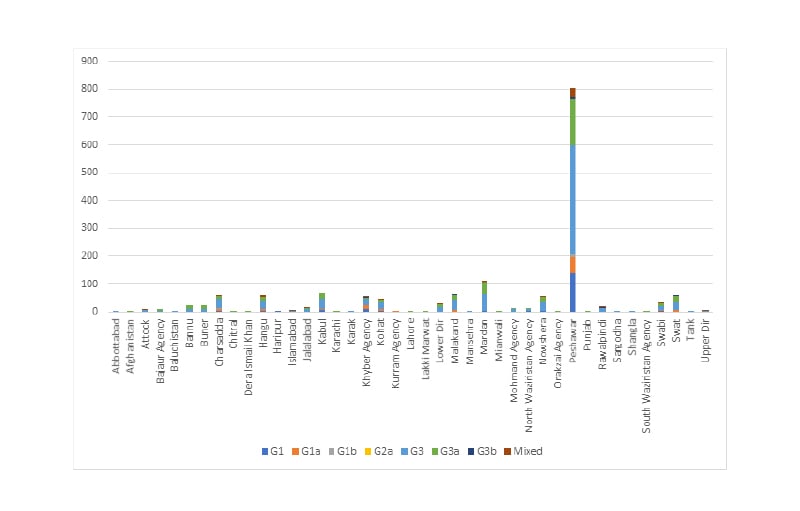
Figure 2: Balance plot of geographic distributions of hepatitis C virus by genotype.
Age- and Sex-Based Distribution of Mixed Genotypes
A total of 39 (3.10%) patients with mixed genotypes were observed. Genotypes 3a+1a were found in 32 patients (82.06%), whereas the 3a+2a genotype was observed in three patients (7.69%), 4a+3a in two patients (5.13%), 3a+1b in one patient (2.56%), and 2a-1a in one patient (2.56%). For the mixed genotypes, 22 (56.41%) were female and 17 (43.59%) were male (Table 1).
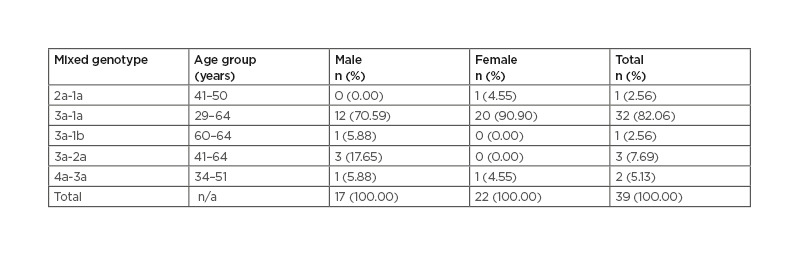
Table 1: Mixed genotype associations with sex and age group in Khyber Pakhtunkhwa, Pakistan. n/a: not applicable.
DISCUSSION
It is now well established that the severity of HCV infection, progression, and patient response to therapy vary according to HCV genotype and subtypes. Worldwide, several regional differences appear to exist in the distribution of HCV subtypes and major genotypes.
Hence, knowledge of the distribution of the various genotypes in Pakistan is essential for the prognostic implications of chronic hepatitis C infection. In this study, the authors reported genotype-based distribution of hepatitis C in relation to age, sex, and geographic locality.
Although several studies have been performed on hepatitis and its genotypes in other parts of the country, a population-based data set is still lacking. It is important to note that Pakistan has the second highest prevalence of hepatitis C in the world next to Egypt; in the Punjab province alone, 7% of individuals are infected with HCV and the rate of infection is still rising.17-19
The prevalence of the various genotypes observed in the study is very similar to those reported in previous studies. A key observation in this study was the finding of the most prevalent genotype, 3a, followed by mixed genotypes, which was also reported by Gul et al.27 However, as reported previously by Idrees et al.26 in a study based in Khyber Pakhtunkhwa, no cases of genotype 5 or 6 were observed in this study.20-25 The most prevalent genotypes in the population of Khyber Pakhtunkhwa was 3a and mixed types, which was also reported by Gul et al.27 The 41–50 years age group was the most infected group, with a slight predominance in female individuals, which has also been previously reported.28-30
The frequency distribution of the HCV genotype observed in this Pakistan-based study appears to be similar to the reported genotypes from other South Asian countries such as India31 and Nepal,32 with 3a as the predominant genotype; however, this pattern differs from the USA, Western Europe,33 Thailand,34 and Japan35 where the reported predominant HCV genotype is 1 (1a and 1b).
CONCLUSION
This study found that the most prevalent HCV genotype in Khyber Pakhtunkhwa, Pakistan, was type 3a followed by 1a, mixed, 1b, 2a, and 3b. This epidemiological pattern of genotype distribution is seen across South East Asia; however, there is no agreed explanation for this observed pattern.20 It was also observed in this study that HCV genotype 3a and mixed genotypes infection is slightly more common in females; on the other hand, males showed slightly higher prevalence of genotypes 1a, 1b, 2a, and 3b. The age group most affected was the 41–50 years age group, followed by the 51–60 years age group. Low prevalence was observed in individuals >70 years of age and <20 years of age. To conclude, all possible modes of HCV transmission should be communicated to the masses through awareness campaigns to control the spread of the virus.