Abstract
The human gut microbiome has garnered much attention over the past two decades with important discoveries linking it to human health and disease. The commensal bacterial flora evolves due to the influence of a number of factors including diet, pathogen exposure, environmental toxicants, disease states, and a challenged microenvironment that requires balancing with the host itself. However, the composition of bacterial species can impact and contribute to the development of local and systemic inflammation. Among the factors attributed to intestinal inflammation are dysbiosis caused by pathogenic bacteria, following decreased host immunity or loss of intestinal barrier function. Dysbiosis can also be triggered by antibiotic therapy or the use of other medications that allow for colonisation of pathogenic bacteria, such as proton pump inhibitors. The imbalance with commensal bacteria leads to the generation of proinflammatory mediators and a reduction of host immune defences, due to a lack of short-chain fatty acid generation needed for energy production to maintain barrier and immune function. The initially localised inflammation results in further dysbiosis as former commensal bacteria are able to breach the barrier and cause systemic immune responses. Low-grade systemic inflammation is a hallmark of inflammatory bowel disease. Because a specific dysbiosis is common in patients with inflammatory bowel disease, it can serve as an early diagnostic marker in its development. Furthermore, faecal microbiome transplants have shown promising benefits in patients with ulcerative colitis and Crohn’s disease.
INTRODUCTION
In the past 10 years, the intestinal microbiome has been recognised as an important contributor to human health and disease. Scientific investigations have focussed on its maturation, interindividual differences, and interplay between the gut microbiome and a broad range of conditions. What was once perceived as a hypothesis with little science to back it up is now a well-regarded fact: the individual microbiome composition influences the host immune system, gastrointestinal health, and interplay with the central nervous system (the so-called ‘gut–brain axis’).1
This does not suggest that the intestinal microbiome is static from birth to death. While the mother primarily provides the initial gut microflora at birth, environmental and interindividual factors impact how the host–bacteria symbiotic interaction develops over time.2 The influence of mode of birth delivery (vaginal versus caesarean) on microbiome composition and development remains controversial. Some research shows no correlation between microbiome diversity and mode of delivery,3 while other trials indicate that specific groups such as preterm deliveries are at greater risk of opportunistic pathogens and stunted microbiota because of the expression of proinflammatory mediators.4 Similarly, babies born via caesarean delivery were more likely to acquire pathogens commonly found in the hospital environment (Enterococcus, Enterobacter, and Klebsiella species), rendering them vulnerable to infections and dysbiosis in early life.5
The various aspects of symbiotic interplay have been established over millions of years as part of evolutionary and adaptive selection among mammals, and remain to be further explored.6,7 In the past decades, the composition of the intestinal microbiome has been analysed to understand correlations between the levels of specific microbiota and the risk for development of chronic disorders such as diabetes,8 obesity,9 inflammatory bowel disease (IBD),10 liver disorders,11 cardiovascular disorders,12 and systemic inflammatory disorders.13 Although the composition of the intestinal microbiome is one contributing factor among many, its contribution may indicate a profound dysbiosis between the human host and detrimental bacterial strains. The complex nature of the microbial composition should be considered when evaluating the individual patient and the nature of their symptoms. For particular disorders, both qualitative and quantitative differentiation among microbial strains that may contribute to the underlying pathophysiology need to be considered.
The diversity of bacterial phylae differs among healthy children and adults with a predominant presence of Firmicutes (approximately 70% and 55%), Bacteroidetes (approximately 12% and 35%), Proteobacteria (approximately 5% and 6%), and Actinobacteria (approximately 5% and 4%) in adults and children, respectively.14 For specific disease states, both the relative quantity of healthy bacterial phyla and invasion of pathogenic strains will contribute to dysbiosis, leading to local and systemic inflammation, disrupted enterocyte barrier function, and host immune responses.15,16 While the phylum Firmicutes are primarily producing the short-chain fatty acid (SCFA) butyrate, and Bacteroidetes producing the SCFA propionate, it is Proteobacteria and Actinobacteria that appear to be primarily involved in health and disease states.17 To date, three predominant healthy microbiome enterotypes have been proposed that can be differentiated by their quantitative composition of Prevotella (genus of Bacteroidetes), Bacteroides (genus of Bacteroidetes), and Ruminococcus (genus of Firmicutes).18
In recent years, the changes in microbiome composition and expression of specific proteins in special populations has been evaluated, indicating a role for the microbiome in the development of such disorders. One population are patients with colorectal cancer that present with abnormal levels of reactive oxygen species, facilitated through increased expression of bacterial superoxide dismutase, leading to oxidative stress in enterocytes and subsequent DNA damage.19 Obesity is associated with the development of colorectal cancer and has also been linked to a dysbiosis of the gut microbiome.20 A mediator for the development of obesity and insulin resistance appears to be the presence of toxic lipopolysaccharides secreted by mostly Gram-negative bacteria.21 Lipopolysaccharides interfere with tight junctions, disrupt intestinal barrier integrity, and can passively diffuse through the enterocyte layer into the systemic circulation to trigger an inflammatory response.22 Similarly, the overabundance in a dysbiotic intestinal microbiome of lipopolysaccharides leads to inflammation of pancreatic islet cells, contributing to the development of diabetes. In addition, a shift in the production of SCFA from propionate to acetate can contribute to reduced production of histone deacetylase inhibitors which are crucial in the regulation of inflammation and glucose sensitivity.
REGULATION OF INTESTINAL AND SYSTEMIC INFLAMMATION
Local intestinal and systemic inflammation are interconnected as blood biomarkers have been elucidated to help diagnose a range of intestinal inflammatory disorders, although additional symptom presentation and differential diagnosis is necessary.23 The epithelial cells of the intestinal lining depend on metabolic products generated by intestinal bacteria, primarily resulting from both carbohydrate and lipid metabolism. Intestinal bacteria metabolise complex and simple carbohydrates and lipids into SCFA, primarily butyric and propionic acid.24 Both are utilised by enterocytes and epithelial intestinal cells as energy sources in primary metabolism (Figure 1).
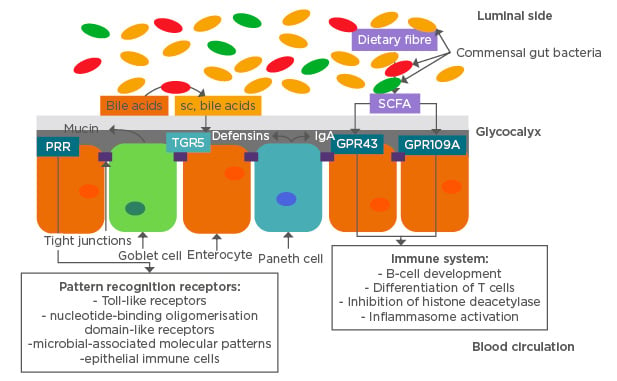
Figure 1: Physical and chemical intestinal barrier functions and their inter-relationship with commensal gut microbiome.
GPR43/109A: G protein-coupled receptor 43/109A; IgA: immunoglobulin A; PRR: pattern recognition receptor; sc: secretory component; SCFA: short-chain fatty acids; TGR5: transmembrane G protein-coupled receptor 5.
The diversity of intestinal bacteria is dominated by the phyla Firmicutes and Bacteroidetes that have been shown to interact with the intestinal barrier function and systemic immune system in a symbiotic manner.2,6 The intestinal bacteria serve as the first defence against environmental disease factors such as viruses, bacteria, and other toxins. In that capacity, the intestinal commensal microbiome is challenged on a daily basis and maintains a highly competitive environment to protect against external bacterial strains.25 Pattern recognition receptors are one component of the link between the intestinal microbiome and the host, and are associated with regulation of immune function.26 Several receptors and immune cells are involved in distinguishing between symbiont and pathogen, and in regulating the microflora, including: toll-like receptors; nucleotide-binding oligomerisation domain-like receptors; microbial-associated molecular patterns, activated by microbial lipopolysaccharides, peptidoglycans, or formylated peptides; and epithelial immune cells (Figure 1).26,27 Primary regulatory pathways utilised by the host to limit proinflammatory responses from the commensal bacteria include nuclear factor κB (NFκB), mitogen-activated protein kinase, and caspase-dependent signalling cascades.26 These pathways lead to induction of apoptosis of pathogenic bacteria and limit the inflammatory response in the surrounding tissue.
A less specific approach to eliminating pathogenic bacteria is the involvement of both the innate and adaptive immune system of the host to release reactive oxygen species, that also leads to the localised loss of symbiotic bacteria.
A major aspect of recognition of commensal bacteria by the host is established through expression of intraepithelial lymphocytes early after colonisation, for differentiation between symbiont and pathogen. Because fucose serves as an energy source for many commensal bacteria, its expression on epithelial glycans favours early colonisation with Firmicutes and Bacteroidetes bacteria and thus helps to establish the early microbiome.28 In order to restrict bacteria from penetrating through the epithelial layer of the intestinal lining, toll-like receptors located in the subepithelial layer and on the basolateral side of enterocytes serve to monitor and activate the immune system.29 Microbial-associated molecular patterns are generated by commensal bacteria to stimulate the host’s innate immune system, provide differentiation from pathogens, activate proinflammatory mediators in the intestinal lumen and the systemic circulation, and recruit T-helper cells to initiate a local inflammatory response.30 Regulated by the adaptive immune system are a range of proinflammatory mediators that both function to maintain the microbiome equilibrium and play a role in the development of inflammatory disorders. The primary proinflammatory mediators involved are interleukins, neutrophils, and stress-mediators (noradrenaline and corticosterone).31-33 This leads to a chronic suppression of commensal microbes in the gastrointestinal tract, which causes damage to mucosal barrier function that triggers an inflammatory and immune response.34
Intestinal barrier functions are maintained by epithelial cells that provide both physical and chemical barriers to protect against invasion of pathogens. At the same time, epithelial cells absorb nutrients and water from the lumen and interact with the microbiome to maintain homeostatic balance.35 The physical barriers separate the gut bacteria from epithelial cells through secretion of a mucus layer, the glycocalyx covering absorptive epithelial cells, and tight junctions linking enterocytes to prevent passage of bacteria and pathogens into the bloodstream. The mucus produced by goblet cells in the large intestine consists of a thick inner and thinner outer layer of O-glycosylated Mucin-2 protein. This protein is utilised by the host in the inner layer to deter bacteria and by bacteria in the outer layer as an energy source after proteolysis to polysaccharides.36 The inner mucus layer is hypothesised to be kept free of bacteria by secretion of IgA and defensin proteins that are part of the chemical barrier (Figure 1). Defensin peptides are antimicrobial peptides secreted by Paneth cells that require activation by matrix-metalloproteinases to protect against Gram-positive and Gram-negative bacteria.37
INTERPLAY BETWEEN MEDICATIONS AND THE INTESTINAL MICROBIOME AND HOST IMMUNE FUNCTION
The commensal microbiome plays a prominent role in both nutrient metabolism and defence against pathogens, and contributes to inter-individual variability in pharmacokinetic and pharmacodynamic effects of drugs. The differences in expression of microbial phyla affect drug metabolism on an individual level. This has been shown for a range of drugs through metabolomics and genomics studies.38 In an analysis of 271 orally administered drugs and 76 different human gut bacteria, the antiviral drug famciclovir was noted to have a significant correlation in altered metabolism in the presence of higher abundance of the bacterial phyla Bacteroidetes and the genera Bacteroides, Parabacteroides, and Alistipes.38 In contrast, the corticosteroid norethisterone was not noted to have significant correlations on both the phylum and genus level. As a consequence, bioavailability of drugs and their respective active metabolites may be altered by individuals’ microbiome, resulting in variable blood levels. Metabolism due to intestinal microbiota is currently given little consideration as a factor in drug bioavailability.
Other factors that can impact the symbiotic relationship between the intestinal microbiome and the host are xenobiotics that disrupt commensal bacterial growth (e.g., antibiotics), and physiological irregularities such as sleep–wake cycle abnormalities.39,40 Antibiotic therapy intermittently disrupts the balance of commensal intestinal bacteria and increases the selection pressure towards potentially pathogenic bacteria. A study explored the expression of antibiotic-resistant genes in commensal bacteria under ciprofloxacin and cotrimoxazole therapy. It found distinct patterns between the two antibiotic therapies, causing either positive or negative selection pressure correlating to higher levels of specific bacterial strains. While ciprofloxacin therapy resulted in higher amounts of Actinomyces meyeri, Acinetobacter johnsonii, and Staphylococcus saccharolyticus, treatment with cotrimoxazole resulted in higher levels of Proteus vulgaris, A. meyeri, and Acinetobacter lwoffi. Ciprofloxacin also resulted in reduction of Citrobacter europaeus, C. koseri, and C. werkmanii, while the yeast Candida tropicalis was less abundant with cotrimoxazole treatment.40 In a separate study with the antibiotic clindamycin using an ex vivo donor-simulated microbiome, a significant reduction in the phylum Bacteroidetes was observed primarily because clindamycin targets primarily anaerobic bacteria.41 Because the Bacteroidetes are significantly contributing to the generation of SCFA such as acetate, propionate, and butyrate, clindamycin antibiotic therapy results in decreased generation and delayed recovery of SCFA synthesis. Given the importance of SCFA to maintain host immune function and intestinal barrier integrity, efforts to restore intestinal microbiome homeostasis as early as possible by administering probiotics or replacing specific mixtures of Bacteroidetes strains are under research.
Dysbiosis may also lead to pathogenic infections including Clostridium difficile, which have been linked with chronic disorders such as metabolic syndrome, IBD, diabetes, and obesity.42 Chronic use of proton pump inhibitors for the treatment of gastric acid reflux disorders has also been shown to reduce the diversity and abundance of the intestinal microbiome.43,44 In two separate clinical trials, the use of proton pump inhibitors led to a significant reduction in bacterial abundance and diversity that may increase the risk of subsequent pathogen infections such as C. difficile. However, the bacterial abundance and diversity was distinctly different from that of C. difficile patients, primarily displaying an increase in Bacteroidetes and Lactobacillales and a reduction in Cyanobacteria and Firmicutes.44
INFLAMMATORY BOWEL DISORDERS AND THE MICROBIOME
The underlying pathogenesis of IBD remains unknown but commonly includes morphological changes in brush border physiology with inflammatory infiltrates, and dysregulation of epithelial barrier function, that often leads to significant loss in quality of life for patients. A common change in intestinal barrier function is reduced expression of defensins and mucin that contributes to a weakened physical and chemical barrier against pathogenic bacteria. With decreased production of the protective mucus layer, even commensal flagellated bacteria such as Escherichia and Proteus species can lead to colitis with subsequent infiltration, due to loss of tight junction integrity.45 The initiation phase leading to loss of barrier functions can be caused by environmental factors or genetic factors, relating to mucosal barrier-related genes including FUT2, MUC19, and NOD2, increasing the susceptibility for development of IBD.35
The localised inflammation is often associated with visceral pain, intestinal motility disturbances, and nutritional malabsorption.46 Many patients present with low-grade elevations in systemic inflammatory markers such as C-reactive protein and white blood cell count.47 These markers are non-specific and further differential diagnosis is necessary, which often makes IBD a diagnosis of exclusion. However, the composition of the microbiome appears to be predictive of a patient’s intestinal inflammatory status. In intestinal germ-free animal models, the introduction of specific bacteria, such as Actinobacteria, Campylobacter, or Enterobacteria, increased the risk for the development of IBD if abundant enough to cause dysbiosis.48,49 Similarly, both ulcerative colitis and Crohn’s disease patients have been noted to have reduced levels of Firmicutes bacteria compared to healthy controls, and Crohn’s disease patients were noted to have lower levels of the anti-inflammatory Roseburia, Phascolarctobacterium, and Faecalibacterium prausnitzii strains.50,51 In addition to dysbiosis of the intestinal bacteria, higher levels of fungi such as Aspergillus clavatus, Candida albicans, and Cryptococcus neoformans have also been detected and associated with the development of Crohn’s disease.52
By further differentiating the generated metabolites of the intestinal bacteria, recent publications indicate that a differential diagnosis is possible.53 A majority of stool samples found an increase in tryptophan and other amino acids and a decrease in SCFA (especially propionic acid). Stool samples were more accurate in differentiating between Crohn’s disease and ulcerative colitis than serum blood or urine samples. SCFA play a crucial role in maintaining intestinal immune barrier function through multiple pathways. They support B-cell development,54 promoting the differentiation and expansion of regulatory T cells through effects on dendritic cells and macrophages,55 maintaining mucosal integrity through inflammasome activation and IL-18 activation,56 and exerting antiproliferative activity through inhibition of histone deacetylase.57 The main receptors for SCFA on enterocytes are GPR43 and GPR109A, and are often dysfunctional or significantly reduced in patients with IBD compared to controls.58 The lower expression of either of these receptors has been proposed as a potential target for treatment of acute inflammation in IBD, but agonists of GPR43 have not been successful to date. In contrast, supplementing SCFA in patients with IBD has shown benefits in reducing acute IBD symptoms.59
Bile acid metabolism is also intricately linked to the intestinal microbiome. Bile acids are deconjugated after aiding in lipid absorption by the enzyme bile acid hydrolase. Bile acids that are not deconjugated are metabolised by bacteria in the colon to secondary bile acids via desulfation that bind to the TGR5 (Figure 1).60 Dysbiosis of intestinal microbiota can lead to a loss of bile acid hydrolase activity thus resulting in less absorption of deconjugated bile acids and more bile acids available in the colon for bacterial metabolism. Disturbed generation of secondary bile acids then influences the signalling cascade mediated through TGR5 leading to increased inflammation and overall lipid malabsorption.61
NOVEL TREATMENT APPROACHES FOR INFLAMMATORY BOWEL DISEASE
The current treatment approaches for IBD emphasise symptomatic relief of pain and the causative inflammation without addressing the underlying pathophysiology. As such, the mainstays of treatment are corticosteroids, anti-TNF-α therapy, and immunosuppressants that are given to suppress the inflammation.62 However, standard therapy options and approaches do not provide adequate relief of symptoms and may often lead to progression in severity of the disease, requiring more aggressive treatment with potential for adverse effects and surgical intervention. Given the complex nature of IBD and the intricate connection between host and microbiome, different approaches have been developed that can be supplemented to alleviate IBD symptoms.
While genetic factors play a role in the development of IBD, they are not utilised in therapy but are primarily used in diagnosis and risk assessment. Environmental factors, however, can be addressed to reduce the risk of development of an IBD. These include diet, smoking, stress, and medications. Diets rich in sugar and long-chain fatty acids accelerate intestinal inflammation and are a known risk factor for the development of Crohn’s disease.46 Smoking increases the risk of developing Crohn’s disease due to its effect on humoral and cellular immune responses, but appears to lower the risk of ulcerative colitis due to the proposed promotion of colonic mucus production. The impact of environmental factors on the progression of IBD is still under research, including the potential of animal proteins for promotion of proinflammatory macrophages leading to colitis.
Microbiome-Focussed Therapies
To alter the intestinal microbiota, two therapeutic approaches are possible. The first approach aims to reduce pathogenic bacteria by using antibiotic therapy, while the second strengthens beneficial bacteria to re-establish symbiosis. Antibiotic therapy has been utilised since the 1970s and since then a range of antibiotic drug classes in combination with immunosuppressants have been used to treat IBD. The greatest rate in remission is observed with the use of rifaximin, a broad-spectrum antibiotic with local intestinal action and low oral bioavailability.63 Metronidazole, either alone or in combination with amoxicillin and tetracycline, has also shown clinical improvement but is not as favourable a therapy due to the possible development of bacterial resistance. Local adverse effects and dysbiosis affecting the overall gut microbiome are the main issues with this approach.
The use of probiotic formulations over the past two decades has gained attention and in recent years specific microbial strains have been developed that lower remission rates in both Crohn’s disease and ulcerative colitis, if taken for a ≥8-week period for ulcerative colitis, and between 10 weeks to 1 year for Crohn’s disease.63 The proprietary probiotic mixture VSL#3 and various Bifidobacteria, as well as the yeast Saccharomyces boulardii, was of benefit to patients with IBD in a majority of clinical studies. Studies not demonstrating a benefit for patients with IBD may be attributable to heterogeneity in doses or protocols. All of the current clinical trial protocols and investigational treatment approaches involving probiotic formulations or faecal transplants are not widely established in clinical practice, or have guidelines for their use, other than the use of Escherichia coli Nissle for maintenance of remission in ulcerative colitis in 5-aminosalicylic acid-intolerant patients.64 An important note on the use of probiotics is the sufficient administration of colony-forming units (CFU) that can reach the intestinal lumen, so need to be formulated accordingly to withstand stomach acid. A minimum of 109 CFU (1,000,000,000 CFU) is recommended per day in order to achieve a therapeutic benefit, which may have to be increased in patients who are concomitantly on antibiotics.
Prebiotics are food items that serve as energy sources for intestinal bacteria and assist in the generation of SCFA, which are found to be low in IBD patients. Supplementation with prebiotics such as psyllium, wheat bran, and oligofructose-enriched inulin resulted in improved clinical outcomes and quality of life but was not always correlated with lower inflammation or remission rates.63 Prebiotics in combination with probiotics (synbiotics) provided a better outcome in regards to remission rate.
The transplantation of faecal microbiota from a healthy donor to a patient with IBD has become a viable possibility with the approval of regulatory agencies to establish live biotherapeutic products and provide guidance on proper safety and handling.63 Faecal microbiota transplantation has gained traction over the past 7 years for patients with severe forms of IBD that do not achieve sufficient symptom relief by any other means. Prior treatment with immunosuppressive and antibiotic drugs is important for successful faecal microbiota transplantation, to avoid native microbiota causing a local inflammatory response. Rectal engraftment in multiple small transfers is generally better tolerated than nasogastric or single-transfer administration. Promising results in maintenance of remission in ulcerative colitis have been found with the use of capsulated freeze-dried donor faecal transplant microbiota that can be taken orally for extended periods of time. In a small open-label study of 30 patients with ulcerative colitis in remission, administration of encapsulated faecal microbiota transplant for 6 weeks did provide adequate therapy without escalation of existing concomitant pharmacotherapy.65 This microbiome-focussed therapy field is still developing, with more controlled clinical trials needed.
CONCLUSION
IBD are complex chronic disorders that remain challenging to diagnose and treat. By gaining a better understanding of the intricate interaction between the host and the intestinal microbiome and its dysregulation in IBD, differential diagnoses and treatment approaches can be better applied in clinical practice. New diagnostic criteria, such as SCFA variations in stool and expressed levels of specific intestinal bacteria, can assist practitioners in confirming a diagnosis and optimising therapy. Such therapies can be tailored to address dysregulation by reducing pathogenic bacteria or increasing beneficial bacteria. Advancing treatment approaches and availability is essential to benefit healthcare providers and patients alike.