Abstract
Background: Escherichia coli is a normal inhabitant of the mammalian gut, but it also exists in a number of pathogenic forms. Diarrhoeagenic E. coli constitute pathotypes that are responsible for fatal infections in humans. Increasing contact between humans and non-human primates provides an opportunity for the transfer of potential pathogens between host species. This study aimed to characterise diarrhoeagenic E. coli pathotypes isolated from baboon faecal samples.
Methods: E. coli were isolated from faecal samples of 124 animals using culture-based methods. Characterisation was achieved by isolating deoxyribonucleic acid from pure isolates and subjecting them to PCR, using specific primers for detection of virulence genes.
Results: Four E. coli pathotypes including enterotoxigenic, enteropathogenic, enterohaemorrhagic and enteroinvasive were detected from baboon faecal samples. The most prevalent pathotypes were enterotoxigenic (14.5%) and atypical enteropathogenic (14.5%) in the captive and the free-ranging populations respectively. However, the observed differences in the pathotype detection were not significant (p>0.05). Enteroaggregative and diffusely adherent E. coli were not detected from any of the study samples.
Conclusion: Olive baboons harbour diarrhoeagenic E. coli pathotypes; hence, the species could be a potential source of zoonotic transmission.
INTRODUCTION
Escherichia coli is a Gram-negative bacilli that exists as the predominant member facultative anaerobic gut microbial flora in humans and other warm-blooded animals.1,2 The colonisation of the human gut by this organism begins shortly after birth, where they remain in the intestinal lumen as commensals throughout their lifetime.3 Together with other gut microbial flora, E. coli confers protection to the host through competitive inhibition of development of pathogens.4 However, some strains of E. coli have evolved by acquisition of virulence, encoding genes through horizontal gene transfer, resulting in highly pathogenic forms of this bacteria that are capable of causing diverse intestinal and extraintestinal infections.5 Strains of E. coli that cause intestinal infections are commonly referred to as diarrhoeagenic E. coli (DEC). These pathotypes cause diarrhoea through different mechanisms, since each possess a unique combination of virulence genes.1 Six strains associated with DEC include: enterotoxigenic E. coli (ETEC), enteropathogenic E. coli (EPEC), enteroinvasive E. coli (EIEC), enterohaemorrhagic E. coli (EHEC), enteroaggregative E. coli (EAEC), and diffusely adherent E. coli (DAEC) have been characterised based on pathognomic virulence factors they produce.6-8
The most common pathotype, particularly in low- and middle-income countries (LMIC), is ETEC. This is due to poor hygiene and inadequate supply of safe drinking water.9 A diverse array of plasmid-encoded colonisation factors are essential for infection. In addition, ETEC produces heat labile and/or heat stable toxins, which are responsible for pathogenesis, culminating in watery diarrhoea that ranges from self-limiting to severe.9,10
The major cause of fatal diarrhoea among infants in LMICs is EPEC, a pathotype that produces attaching and effacing (A/E) lesions in the intestinal mucosa.11,12 Human infections with EPEC are marked with invasion of the colon and disruption of tight junctions A/E lesions on the microvilli, leading to watery diarrhoea.13 Formation of A/E lesions is regulated by locus enterocyte effacement that constitutes a pathogenicity island populated with virulence genes, including eae that encodes for intimin.14 Typical EPEC (tEPEC) are characterised by the presence of the bfp gene that encodes for adherence factors; this is absent in the atypical (aEPEC) sub-pathotype,15 and forms the basis of genomic characterisation.16
EIEC cause bacillary dysentery of a lesser severity compared with Shigella spp. Like Shigella spp., EIEC is endocytosed by microfold cells of the follicle-associated epithelium to access the colon submucosa.5,17 Pyroptosis of macrophages that have phagocytosed EIEC release the free organisms that invade enterocytes, leading to their intracellular replication and dissemination to other cells.17 In addition, pyroptosis induces inflammatory response capable of destroying the infected epithelium.18
Most outbreaks of severe diarrhoea in adults and children are caused by EHEC, which is harboured by cattle as a reservoir.11 It is highly infectious and produces Shiga-like toxins (STEC) that belong to the Stx family: Stx1 and Stx2. Like EPEC, it also possesses a locus enterocyte effacement pathogenicity island that is responsible for the A/E lesions in the microvilli produced by both pathogens.19 Another important bacterial cause of travellers’ diarrhoea is EAEC.20 This infection is characterised by persistent diarrhoea among malnourished children, and this pathotype has been ranked as the second most commonly isolated bacteria in patients with travellers’ diarrhoea.21,22 It forms a characteristic brick-like aggregative adherence when grown in HeLa cell cultures.23 Virulence factors associated with EAEC include aggregative adherence fimbriae, heat stable enterotoxin, and haemolysin E to colonise intestinal epithelium, stimulate mucus secretion, and trigger cytotoxic mucosal damage and inflammation, in addition to formation of biofilms.24
Diffusely adherent E. coli form diffuse adherence to HeLa or Hep-2 cell lines in vitro, since they produce adhesins encoded by Afa/Dr operons.25 This strain is responsible for diarrhoea among children, with its incidence increasing from the age of less than 5 years.26
Despite the role of DECs as enteric pathogens responsible for fatal infections in humans, they have been isolated from different non-human primates (NHP) except baboons.27 Considering that there is increased contact between humans and NHPs due to different anthropogenic activities such as farming, fragmentation, and logging,28,29 presence of DECs in these hosts poses a high risk of zoonotic transmission that could result in escalated morbidity and mortality.30 Some pathotypes have been reported in asymptomatic NHPs, including marmosets, tamarins, white faced sakis, De Brazza’s, and spider monkeys, but not in baboons.27,31 In this study, the authors examined E. coli isolates from 124 olive baboons (Papio anubis). Pathotype proliferation in NHPs is a potential threat to human health. An enhanced understanding of existence of pathotypes in NHPs will be critical, not only to maximise and maintain human health, but also to shape the understanding of disease, and to foster new preventive and therapeutic approaches.32 Strategies to limit transmission of pathogens between human and NHPs would benefit both human and primate conservation.33 The goal of this work was to characterise P. anubis DECs isolated from captive and free-ranging P. anubis and to assess their potential as a source of infection to humans, in order to mitigate the risk of zoonotic infections.
METHODS
Study Animals
This study involved two equal groups of P. anubis (n=62) that represented both captive and wild populations. Free-ranging P. anubis within the Mpala Research Centre, Laikipia County, Kenya, constituted the wild population. The centre contains over 25 wild mammalian species, including baboons, that interact closely with pastoral communities and their livestock. On the other hand, captive P. anubis were housed at the Institute of Primate Research (IPR), Nairobi, Kenya, in standard animal enclosures. This is a World Health Organization (WHO) collaborating centre, accredited by the Association for Assessment and Accreditation of Laboratory Animal Care (AAALAC), which ethically utilises laboratory animals, including NHPs, as guided by national and international standards for animal care and use. All captive baboons were fed on commercial monkey cubes (Unga Farm Care Limited, Nairobi, Kenya), and supplemented with fruits, vegetables, and water ad libitum. This study was approved by IPR’s Institutional Ethical and Research Committee (ISERC; Ref: ISERC 12/16).
Collection of Faecal Samples
Fresh faecal samples were collected from both captive and wild P. anubis by carefully rolling sterile cotton tipped swabs on the surface of freshly voided faecal matter, while avoiding contaminated portions from the ground. The free-ranging animals were carefully followed to ensure that samples were collected from different baboons. These swabs were immediately inoculated into a transport medium (Stuart Transport Medium, Oxoid, Basingstoke, UK), packaged in standard triple package, and transported to microbiology laboratories at IPR for analysis.
Bacteria Isolation and Identification
Target bacteria E. coli were isolated by inoculating the faecal samples onto MacConkey and xylose lysine deoxycholate agar, then incubating the plates at 37 oC for 24 hours. Colonies that fermented lactose were picked and sub-cultured onto MacConkey agar plates and incubated for another 24 hours in order to obtain pure lactose fermenting colonies, which were presumptively identified as E. coli. Identification and confirmation of E. coli was performed using Gram staining reaction, biochemical tests based on analytical profile index assays for enterobacteriaceae (Analytical Profile Index 20E), and oxidase tests as previously described.34
Isolation of DNA
Pure E. coli isolates were inoculated onto tryptone soy agar and incubated for 24 hours at 37 oC. Several colonies were carefully picked using a sterile cotton-tipped applicator stick and suspended in 500 μL of nuclease free water in an Eppendorf tube (Eppendorf, Hamburg, Germany). This suspension of E. coli isolates was boiled using a heat block at 95 oC for 10 minutes, then chilled on ice. This was followed by centrifugation at 10,000x g for 5 minutes in order to pellet cell debris and leave isolated DNA as clear supernatant; this was transferred into nuclease free Eppendorf tubes and stored at -20 oC, awaiting downstream assays.35,36
PCR Assays
Isolated DNA of the E. coli isolates were amplified in 0.2 ml PCR tubes containing 10 μL 5X PCR buffers, 0.2 mM deoxyribose nucleotide triphosphate s mixture, 2.5 U Taq DNA polymerase, 0.15 μmol of each primer, and 1 μL template DNA to make a total reaction volume of 20 μL. DNA templates from known E. coli pathotypes were used as positive controls during the assay. Primers listed in Table 1 were used in this study to target elt and/or est for ETEC, eae and bfpA for EPEC, stx1 and 2 for EHEC/STEC, ipaH for EIEC, CVD432 for EAEC, and daaC for DAEC. Thermocycling conditions for all reactions for each primer involved an initial denaturation step of 5 minutes at 95 oC, followed by 30 cycles of 20 seconds denaturation at 95 oC, 20 seconds annealing at temperatures that varied depending on each primer requirements (Table 1), and 30 seconds extension at 72 oC, followed by a final extension for 3 minutes at 72 oC. Positive controls that consisted of DNA isolated from E. coli (ATCC 43887, ATCC 933J, ATCC 933W, and EAEC PC-strain 17-2 [(O3:H2]) were processed using conditions similar to test samples and the negative control (E. coli microbiology hub-Kericho 0028).41
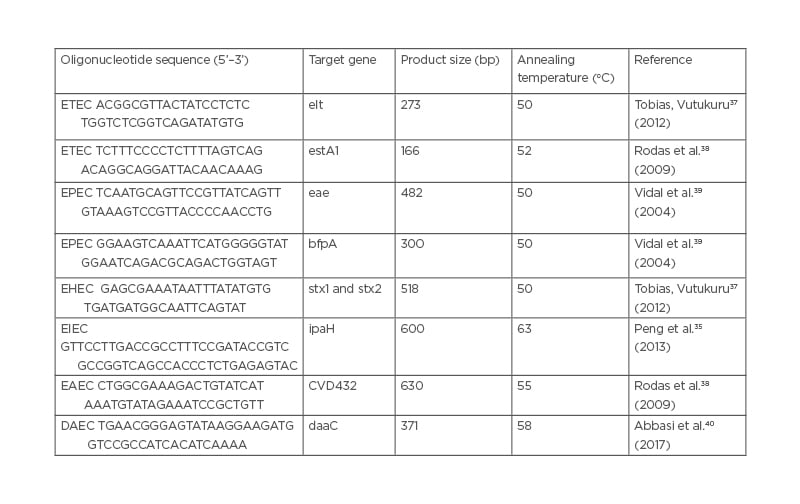
Table 1: Primer sequences used for characterisation of Escherichia coli pathotypes.
Amplified PCR products were subjected to electrophoresis on 1.5% agarose gel, then visualised using the ultraviolet transilluminator documentation system (UVP Bio-Doc It™ Imaging System, Upland, California, USA). This involved mixing 10 μL of PCR products, including the positive and negative controls, with the loading dye, loading carefully into the wells of pre-cast 1.5% agarose gel placed in an electrophoresis tank. A 100 bp molecular marker was put in wells on both ends of the agarose gels in order to determine the size of the amplicons. The electrophoresis tank was connected to the power supply and allowed to run for 1 hour. The agarose gel was then removed from the tank carefully and placed on a UV transilluminator for visualisation. The amplicons’ sizes were then determined and recorded.
RESULTS
Four pathotypes that included ETEC, EPEC, EIEC, and EHEC were isolated from faecal samples from both groups of baboons. The most dominant pathotype among captive baboons was ETEC (14.5%), followed by EHEC (3.2%), aEPEC (3.2%), tEPEC (1.6%), and EIEC (1.6% [Table 2]). On the other hand, aEPEC (14.5%) appeared to be most prevalent pathotype, followed by tEPEC (8.1%), EIEC (3.2%), and ETEC (1.6%), whereas EHEC was not isolated in any of the wild baboon faecal samples. However, EAEC and DAEC were not isolated from any faecal sample from baboons in either of the two groups. The overall observed differences in the isolated pathotypes were not significant (p>0.05).
PCR Products
Confirmed E. coli isolates were evaluated for presence of pathotype-specific virulence genes using PCR assays and visualised under an ultraviolet transilluminator. Amplicon sizes of PCR products were determined against 100 bp molecular marker. Virulence genes representing the four DEC pathotypes detected in this study included elt for ETEC (Figure 1A), eae and bfp genes for EPEC (Figure 1B), ipaH for EIEC (Figure 1C), and stx1 and 2 for EHEC (Figure 1D).
DISCUSSION
The gastrointestinal tract is well known to harbour E. coli, as a common commensal in humans and animals. While E. coli is a normal inhabitant of the mammalian gut, it also exists in a number of pathogenic forms or pathotypes, including those with predisposition for the gastrointestinal tract.This study established presence of four E. coli pathotypes; ETEC ( Figure 1A ), EPEC ([h1]Figure 1B[/hl]), EIEC (Figure 1C), and EHEC (Figure 1D) in faecal samples of P. anubis. As previously reported in other NHP studies,27,30,31 these results have demonstrated presence of E. coli pathotypes in P. anubis. The most prevalent pathotypes were ETEC (14.5%) and aEPEC (14.5%) in captive and free-ranging groups respectively (Table 2). However, the overall observed differences in the isolated pathotypes were not significant (p>0.05).
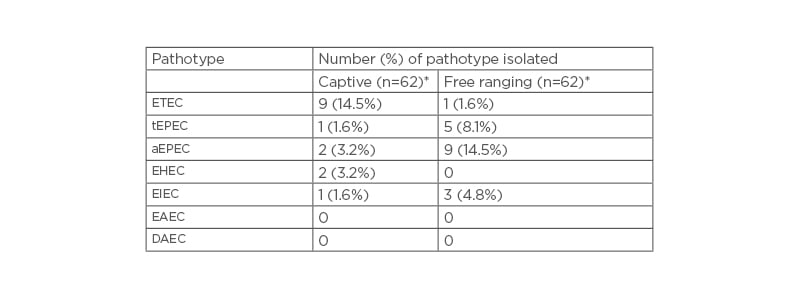
Table 2: Escherichia coli pathotypes isolated from captive and wild olive baboon faecal samples.
*n=62; number of baboons per group.
AEPEC: atypical enteropathogenic Escherichia coli; DAEC: Diffusely adherent Escherichia coli; EAEC: Enteroaggregative Escherichia coli; EHEC: enterohaemorrhagic Escherichia coli; EIEC: enteroinvasive Escherichia coli; ETEC: enterotoxigenic Escherichia coli; tEPEC: typical enteropathogenic Escherichia coli.
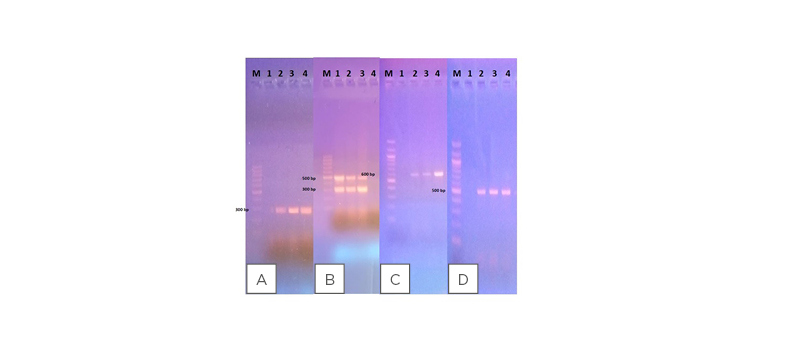
Figure 1: Agarose gel electrophoresis showing amplified PCR product sizes against 100 bp molecular marker (M).
A) The elt gene associated with ETEC: 1 is the negative control; 2 is the positive control; and 3 and 4 are samples that are positive for ETEC. B) EPEC virulence genes (eae and bfp): 1 is the positive control; 2 and 3 are samples that are positive for EPEC; and 4 is the negative control. C) The ipaH gene detected in EIEC isolates: 1 is the negative control; 2 is the positive control; and 3 and 4 are samples that are positive for EIEC. D) The stx 1 and 2 genes associated with EHEC: 1 is the negative control; 2 is the positive control; and 3 and 4 are samples that are positive for EHEC.
EHEC: enterohaemorrhagic Escherichia coli; EIEC: enteroinvasive Escherichia coli; EPEC: enteropathogenic Escherichia coli; ETEC: enterotoxigenic Escherichia coli.
The observed difference in the prevalence where three pathotypes: ETEC, EPEC, and EIEC, were detected in free-ranging P. anubis faecal samples, whereas four pathotypes: ETEC, EPEC, EIEC, and EHEC, were isolated from the captive group, and could be attributed to variations in the environmental settings of the animal populations sampled. The captive baboons are socially housed within IPR with stringent husbandry conditions, where they routinely closely interact with animal healthcare personnel; thus, there is a high possibility of transmission of these pathotypes during cleaning or feeding.42 Routine introduction of environment enrichment material could also be an important source of infection among the captive baboons. Detection of E. coli virulence genes in wells and boreholes has been reported,43 thus implicating them as a potential source of infection to free-ranging baboons, since this important resource is shared by both humans and animals in most African settings. In addition, free-ranging baboons consist of troops that are frequently in close contact with humans, livestock, and other feral animals within the region that increase their risk of transmission, including E. coli pathotypes.28 Presence of EPEC, EHEC, and EIEC have been reported in previous studies to be associated with other NHPs, including De Brazza’s, spider monkeys, white-faced sakis, lemurs, and tamarins.27 Bacterial transmission among humans, mountain gorillas, and livestock has also been reported in some studies, thus establishing that interaction between these species propel zoonotic transmission.44
The four pathotypes ETEC, EPEC, EIEC, and EHEC that were detected in faecal samples from the study baboons have also been documented to be present in faecal samples collected from healthy NHPs in a zoo setting.27 This implies that NHPs can be an important source of DEC infection to humans who come into contact with their faecal materials by virtue of occupation or anthropogenic activities. Presence of ETEC and STEC/EHEC have been demonstrated in cattle manure and faeces,45 implying that bovine waste could be a potential source to the wild infections reported in this current study, considering that Mpala Research Centre is also a grazing zone for pastoral communities.
Although all baboons harbouring E. coli pathotypes were asymptomatic, fatal outbreaks of diarrhoea caused by EIEC have been reported in rhesus macaques and another by EHEC in cynomolgus macaques.31 Presence of these microbes in baboons and other NHPs, both in captivity and in free-ranging groups, is not uncommon. Generally, it has proven difficult to eliminate the most infectious pathogens from these laboratory animals, even when different approaches that work in other species have been employed.46 This is partly attributed to the fact that unlike most laboratory animals, NHPs require to be socially housed in highly-enriched enclosures similar to their natural habitat, as a regulatory requirement for animal welfare and breeding purposes,47 a practice that promotesthe introduction and transmission of pathogens. Typical EPEC-producing characteristic A/E lesions have been demonstrated in both symptomatic and asymptomatic marmosets.48 However, there have been reported cases of haemorrhagic diarrhoea outbreaks in marmosets, opportunistic infections among AIDS-infected rhesus macaques, and ulcerative colitis in cotton-top tamarins.49,50 In China, aEPEC have been isolated from golden snub-nosed monkeys that had diarrhoea.51 These pathotypes have been demonstrated to cause fatal outbreaks in humans,5,52,53 thus presenting them as important zoonoses. Prevalence of diarrhoeal illnesses caused by aEPEC is higher than that of tEPEC among children in LMIC.16,53,54 As an emerging DEC, aEPEC has been detected in foods of animal origin, and in patients from both settings.55,56,57 Infection by ETEC is most prevalent in these regions, with its effects graver among children.9,58 None of the study animals from both groups showed signs of diarrhoea. Interactions of humans with P. anubis that harbour these pathotypes could result in transmission of DEC. As previously reported in other NHPs,48 the current study confirms that P. anubis are potential reservoirs of DECs.
CONCLUSION
Increasing contact between humans and NHPs provides an opportunity for the transfer of potential pathogens between host species. Presence of these pathotypes in the P. anubis species could represent the underestimated source of zoonotic transmission, resulting in high morbidity and mortality. This study provides insight into the disease transmission process, enabling for the calculation of potential risk factors associated with human encroachment on wildlife populations, and vice versa.







