Abstract
Renal diseases are consecutive to a deregulation of gene expression regulated by non-coding RNAs. These non-coding RNAs were discovered at the turn of the 21st century when it was established that post-transcriptional regulation was performed through small non-coding RNAs, known as microRNAs (miRNAs). Up to 3,000 miRNAs are expressed by human cells. They are small, single-stranded nucleic acids, which trigger translational repression of mRNA by base-pairing with the 3′ untranslated region of their mRNA targets. In addition to miRNA regulation, it was also demonstrated that 60,000 long non-coding RNAs are expressed in the human cell and that they are able to regulate gene expression at all levels. The roles of these various RNA families are just beginning to be understood in the field of nephrology. In the past decade, the authors and various others have published that several miRNAs are deregulated during the onset of chronic kidney disease (CKD) and are associated with cardiovascular damage. This review focuses on miRNA-223 (miR-223) as its expression is increased in vivo in the large vessels of a mouse model of CKD, whereas it is diminished in the serum of both mice and human patients with CKD. In patients, miR-223 expression was correlated with all-cause mortality, as well as cardiovascular and renal events. Molecular clues were given by a multi-omics approach, indicating that miR-223 modulates gene regulation at all levels including mRNA expression, protein amounts, and metabolic molecule accumulation. miR-223 is thus a potential target to prevent or treat complications of CKD pathogenesis.
INTRODUCTION
RNAs are currently a much-discussed molecular species. They can be used as vaccines, as extensively shown during the COVID-19 crisis,1 but could also be at the very origin of life on our planet. Indeed, Walter Gilbert coined the term ‘RNA world’ in 1986 to suggest that self-replicating RNA molecules were at the very origin of life during evolution, before the appearance of DNA and proteins.2,3 These RNAs, several billions of years later, would be at the origin of the increasing number of RNA species that are instrumental in gene regulation.4 Short non-coding RNA species such as microRNAs (miRNAs) were discovered at the onset of the 21st century.5,6 The recent progress in deep sequencing has now shown that, contrary to what most molecular biology books describe, a vast majority of the eukaryotic genome is transcribed, and that a complex network of long and short transcripts, including tens of thousands of non-coding RNAs, is central to our metabolism. These findings extend far beyond the DNA–RNA–protein central dogma published by Francis Crick in 1957.7 All these non-coding RNAs are increasingly studied by the authors and numerous others as innovative biomarkers but also as innovative therapeutic targets.
THE ROLES OF NON-CODING RNA IN THE FIELD OF NEPHROLOGY
Patients with later-stage chronic kidney disease (CKD) exhibit a high cardiovascular morbimortality associated with cardiovascular disease (CVD).8 CVDs account for 30% of all global deaths and are particularly devastating in the course of CKD.9 A majority of fatalities are caused by atherosclerosis, a pathological process that will impact any artery. Due to the intricacy of the pathophysiological process, it is thought that non-traditional predictive factors for CVD complications would be useful for prognosis and diagnosis.
miRNAs represent a new class of biomolecules, endogenous interfering RNAs, which are now considered promising biomarkers for numerous pathologies.5 They are non-coding RNAs that contain approximately 20–25 nucleotides and precisely regulate gene expression, thereby affecting the stability and translation of mRNAs in a post-transcriptional manner.5 Their number is now estimated to be approximately 2,000–3,000 and the reciprocal genes represent approximately 3% of the genome.5,10 miRNA precursors are derived from longer primary transcripts termed pri-miRNAs. These are cut in the nucleus by the Class 2 ribonuclease (RNase) III enzyme Drosha into a precursor miRNA hairpin (60–70 nucleotides). The hairpin then migrates into the cytoplasm by way of exportin 5. Dicer RNase III subsequently cuts the hairpin it into a double-stranded RNA, comprising the 5’ and 3’ strands, and will in turn bind the RNA-induced silencing complex. This multi-molecular complex unwinds the RNA into single-stranded RNA, known as the mature miRNA, and directs it to the target mRNAs, which triggers gene silencing.5 Human miRNAs mostly act by destabilising their target mRNAs to diminish translation levels.6 It is now documented that the 2,000–3,000 miRNAs alter the expression of approximately one-third to two-thirds of the human genome.6 This is explained by the fact that mismatches are present in the binding sites between miRNA and mRNA targets and, therefore, a miRNA can bind dozens of different messenger RNAs and attenuate the expression of as many genes.
Since their discovery, it has been understood that miRNAs could be useful tissular biomarkers, however, their use was precluded for most diseases because biopsies were not accessible to biologists. Fortunately, since 2008 we have known that miRNAs also circulate and are stable in human blood.11,12 This makes them useful as non-invasive biomarkers as a simple blood sample is enough to assess their expression,10 and several studies in the field of oncology have shown a link between miRNA seric levels and tissue amount.11 miRNAs are present in microvesicles or complexed with chaperone proteins that protect them from RNase activity,13 emphasising their roles as potential non-invasive biomarkers. A pioneering study from De Rosa et al.14 showed that miRNA levels are different in the serum of patients with coronary artery diseases compared to patients who are healthy. An accurate amount of a spike-in exogenous, synthetic Caenorhabditis elegans miRNA-39 miRNA is mostly used as reference in order to avoid further experimental bias.12
THE IMPORTANCE OF MICRORNA-223 IN CHRONIC KIDNEY DISEASE
CKD is characterised by a slow and progressive loss of kidney function, leading to vessel and bone damage. It has been shown that miRNAs are involved in the pathophysiology of CKD;10,15,16 for example, they play an important role in controlling vascular smooth muscle cell trans-differentiation during vascular calcification, which is a complication of CKD.16,17 This would imply that miRNAs are critical players in the differentiation of vascular cells in CKD-related CVD.
The authors were the first group to show that miR-223 expression levels were modulated in pathological vessels and blood of a CKD mouse model.17,18 In the aorta, miR-223 levels were doubled in the mice with CKD, whereas they almost tripled in ApoE-knockout rodents that suffered from CKD, displaying vascular calcification.17 Conversely, the authors showed that miR-223 seric levels diminished during CKD in all pathological mice;17 this finding was confirmed in a large CKD patient cohort, which will later be discussed.19 Additionally, in the murine study, the authors published that the calcium-free phosphate binder sevelamer carbonate alleviated the miRNA deregulations in aortas, suggesting a possible direct link between the observed miRNA alterations and vascular calcification formation.8 This is interesting in light of miR-223’s role in osteoclast differentiation, suggesting that this RNA has an effect on the vascular calcification and osteoporosis process.20
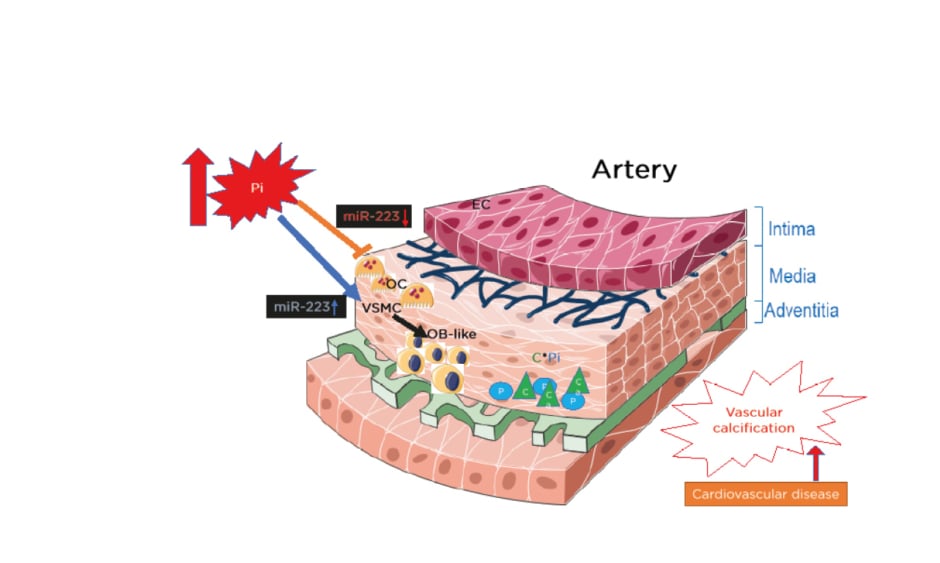
In view of miR-223 expression in the media of the aorta, the authors also observed the expression in the intima (Figure 1). In the intima, miR-223 is strongly expressed in freshly isolated endothelial cells but its levels decrease very quickly during cell subculture.21 This suggests that miR-223 is not produced in endothelial cells but is imported from other cell types, such as monocytes and macrophages. Tabet et al.22 also showed that high-density lipoproteins can deliver miR-223 to endothelial cells. Additionally, the authors published that miR-223 is expressed in freshly isolated endothelial cells from the brain microvasculature.18 The important increase of the inflammatory miR-223 expression during the course of CKD in both the intima and media could thus induce chronic low-grade inflammation and atherosclerosis. Recent results from the authors in a rat model of restenosis suggest that an increase in miR-223 is detrimental and that an anti-miR antisense strategy against miR-223 decreases restenosis (Figure 2).23
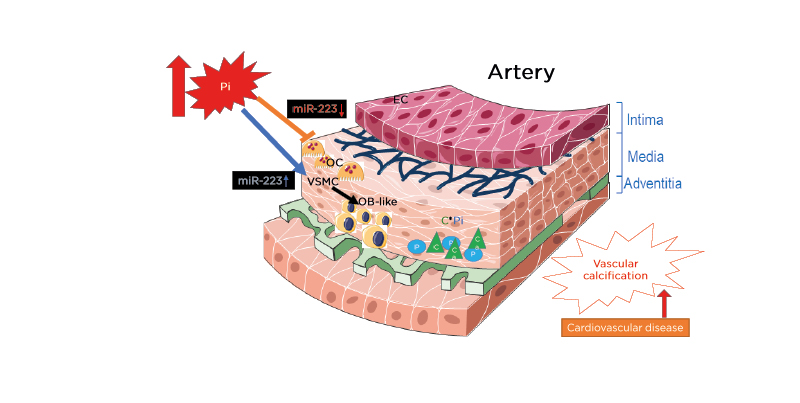
Figure 1: The instrumental role that miR-223 plays on the effects of inorganic phosphate on both VSMC trans-differentiation and vessel osteoclastogenesis, inducing vascular calcification.
C: calcium; EC: endothelial cells; miR-223: microRNA-223; OB: osteoblasts; OC: osteoclasts; P: phosphate; Pi: inorganic phosphate; VSMC: vascular smooth muscle cells.
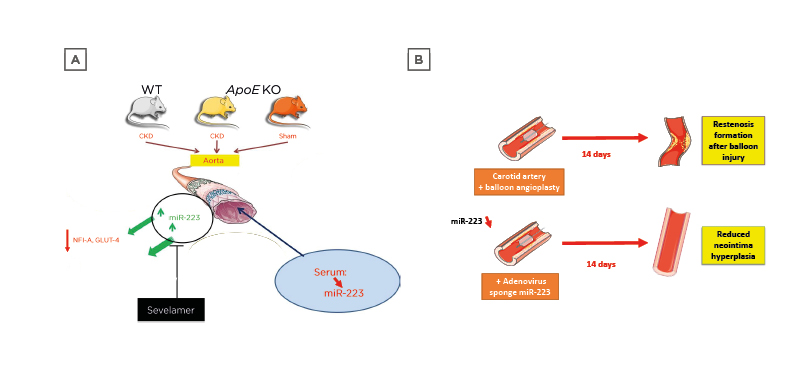
Figure 2: Pre-clinical models explaining the effects of microRNA-223 in vivo on cardiovascular events linked to vascular calcification.
A) Murine model of CKD and atherosclerosis (ApoE knockout). Mice with both CKD and ApoE knockout display vascular calcifications. B) Rat restenosis model. Sponge treatment decreases miRNA-223 levels and diminishes restenosis in the carotid. CKD: chronic kidney disease; GLUT-4: glucose transporter Type 4; KO: knockout; miR-223: microRNA-223; NFI-A: nuclear factor I A-type; WT: wild type.
MICRORNA-223 AFFECTS SEVERAL LEVELS OF GENE REGULATION
A multi-omics study enabled the authors to estimate the influence of over-expression and inhibition of miR-223, a pleiotropic regulator of metabolic-related disease, in a monocyte-macrophage cell line.24 Using a combination of microarray transcriptomics, SELDI-TOF and MALDI-TOF proteomics, and nuclear magnetic resonance-based metabolomics, the authors estimated the levels of proteins, mRNAs, and metabolites to identify genes involved in miR-223 regulation, hypothesising that the deregulated molecules would be candidate disease biomarkers and potential therapeutic targets. They found that 52 proteins were significantly altered when comparing scramble and pre- and anti-miR223 treatment. Among these, the transcription factors CARM1, UBE2G2, Cactin, and NDUFAF4 were involved in the stability of mRNAs, bone remodelling, and immune response. Transcriptomics showed changes in the expression of 120 genes, among which 30 genes encoded for long non-coding RNAs. Representative genes were again implicated in bone remodelling and RNA regulation. The most deregulated metabolites were linked to metabolism and pyrimidine nucleotides.
MICRORNA-223 CAN BE A PREDICTOR OF CHRONIC KIDNEY DISEASE PROGRESSION
Levels of several miRNAs could be associated with CKD progression;10 however, their association with clinical outcomes remained only partly understood. To gain further insight, the authors recently measured serum levels of miR-223 and miR-126 in a cohort of 628 patients (patients with CKD Stage 1–5 or patients on renal replacement therapy [CKD 5D], and healthy controls) in collaboration with the Ghent University Hospital, Belgium. In this large cohort, patients with CKD were followed over 6 years, tracking all-cause mortality and cardiovascular and renal events.19 The serum levels of miR-223 were significantly lower in the CKD Stage 3B–CKD 5D groups than in the control group. Seric levels of miR-126 were significantly lower in the CKD Stage 2–CKD 5D groups than in the controls. Concerning events were that patients with below-median levels of miR-223 and miR-126 had the lowest survival rate. CKD is thus associated with a decrease in circulating miR-223 and miR-126 levels in humans with impaired kidney function. Neither miR-223 nor miR-126 was a prognostic marker of all-cause mortality, cardiovascular events, or renal events, as associations between miRNA levels and overall mortality were not significant when results were adjusted for baseline estimated glomerular filtration rate. The mechanism that underlies the low expression of circulating miRNAs in CKD has yet to be identified.
There are thus several limitations to the use of miRNAs as biomarkers in the area of CKD. Currently, they are no better than the estimated glomerular filtration rate gold standard and, therefore, other candidates will have to be identified. They are also not tissue-specific as miR-223 is expressed in skeletal muscle25 as well as the kidney.
CONCLUSION
miRNA regulation during the course of CKD is a complex process as these small non-coding RNAs are expressed in a cell- and tissue-specific manner. Additionally, the existence of long non-coding RNAs has reshaped our view on gene regulation, increasing its complexity even further. We are now at a stage where non-coding RNA expression can be studied in human CKD populations, before and after the appearance of vascular disease, in order to develop them as new biomarkers, useful for diagnosis and treatment evaluation, but also to detect innovative targets for future therapeutic strategies.