BACKGROUND AND AIMS
Gene expression profiling has a significant impact on understanding the biology of breast cancer (BC). Over the past 15 years, four main intrinsic molecular subtypes of BC have been identified.1-3 At the 13th Saint Gallen International Breast Cancer Consensus, a surrogate classification of BC molecular subtypes by immunohistochemistry (IHC) was established.4 The most controversial point was the difference between the luminal A and luminal B subtypes according to the Ki67 values.5 Commonly, 14% is the Ki67 cut-off that has been established for differentiating BC subtypes; however, in later studies this value has been questioned and a cut-off of 20% has been proposed.6 This study aimed to analyse the correlation between the surrogate BC subtypes using IHC and PAM50 gene expression assay,7-10 considering Ki67 as an independent factor to identify the best Ki67 cut-off.
MATERIALS AND METHODS
Included in the study were females diagnosed between 2015 and 2020 with early stage luminal BC whose samples underwent genomic testing using PAM50/Prosigna® (NanoString Technologies, Seattle, Washington, USA). A total of 143 samples were analysed at a single institution. The IHC subtypes were classified using two independent Ki67 cut-offs, 14% and 20%, and these were compared to the subtypes identified by PAM50.
RESULTS
Using the Ki67 cut-off >14% (Figure 1A), a correlation of 70.6% with a sensitivity of 79.1% and a specificity of 55.8% was observed, as well as a positive predictive value of 75.8% and a negative predictive value of 60.4%. By modifying the Ki67 cut-off to be >20% (Figure 1B), the percentage of well-classified tumours as determined by IHC was 76.2%, improving the agreement by 6.2%. The sensitivity was 93.4%, but the specificity was 46.1%. The positive predictive value was 75.2% and the negative predictive value was 80.0%, which suggests that IHC has a high probability of diagnosing luminal A and luminal B.
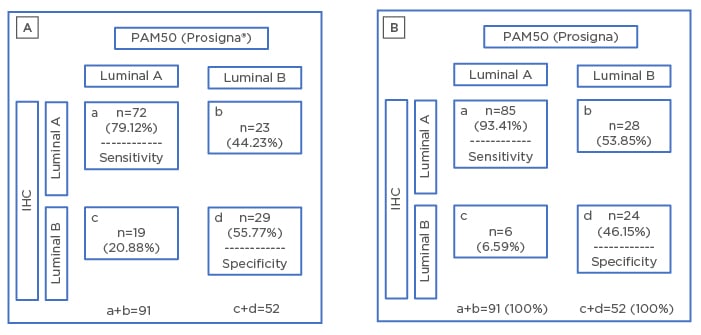
Figure 1: Concordance between immunohistochemistry and PAM50 using two different cut-off values.
A) Concordance between IHC (Ki67 cut-off 14%) and PAM50. B) Concordance between IHC (Ki67 cut-off 20%) and PAM50.
IHC: immunohistochemistry.
CONCLUSIONS
Based on the results from this study, the authors demonstrated that modifying the Ki67 cut-off to >20% provides a better surrogate classification by IHC and a higher sensitivity for classifying the luminal subtypes than ≥14%. The authors propose that the Ki67 cut-off should be globally modified to >20% as an independent factor; however, due to the low specificity, other factors, such as progesterone receptor expression, should be considered.








