Abstract
The cancer burden continues to grow with enormous physical, emotional, and financial pressure on individuals, families, communities, and health systems. Early detection and effective treatment are crucial. The analysis of epigenetic biomarkers is presented as an exceptional solution for early cancer diagnosis and personalised treatment design. These brand new biomarkers have initiated a diagnostic revolution because of their predictive capability and reversibility, opening the window for timely diagnostics and personalised medicine. In recent years, the potential of optical biosensors for epigenetic biomarker evaluation has been revealed. Nanotechnology is promoting the appearance of new advanced biosensors able to be integrated in complete lab-on-chip platforms. Lab-on-chip biosensors are offering simplified, cost-effective, and fast results to solve the current diagnostic problems. In this review, we present the advantages offered by the analysis of epigenetic routes in cancer diagnosis and the current advances in optical biosensors for cancer epigenetic analysis, showing how the new biosensor solutions manage to surpass the challenges encountered during the analysis of each epigenetic mechanism.
INTRODUCTION
Cancer is one of the leading life-threatening diseases across the world, with 200 different types of cancer accounting for >1,500 deaths per day.1 The main constraints in cancer management can be found in two principal problems: late diagnosis and low efficiency of therapies. Despite the recent technological advancements, cancer diagnosis is still an unfinished issue for physicians, since many cancers remain asymptomatic until relatively late stages, leading to a poor survival rate.1 The continuous failure of many treatments against late-diagnosed cancers is forcing scientists to establish new formulas for cancer prediction and staging. A better definition of the tumour origin and its stage of development will reduce the possibility of treatment defeat. Current conventional methods are inefficient for early stage cancer detection, as they are based on the evaluation of the phenotypic properties of the tumour.1 In addition, these techniques are invasive, expensive, and rely on time-consuming procedures carried out in specialised laboratories. Regarding the applied therapies, most of them are not as precise and accurate as required, resulting in low success rates or devastating side-effects, which makes the identification of more specific drugs an indispensable requisite, and proper doses that best benefit each individual.
There are several molecules that express prominent alterations in their expression during cancer, recognised as biomarkers and having high clinical significance. The presence, absence, or change in the level of these specific biomarkers in a cell, tissue, or biofluid often indicates cancer development.2,3 Cancer is a multistage disease of which onset and progression are associated with a complex variety of genetic or epigenetic alterations, resulting in tumourigenic transformation and progression.4 Epigenetics literally means ‘above’ or ‘on top of’ genetics. It refers to any heritable trait that is not derived from the DNA sequence. These inherited characteristics are transmitted to offspring in the form of subtle chemical modifications to DNA and DNA-associated proteins, and exert their effects by modulating the gene expression. Epigenetic mechanisms are implicated in several processes, playing important roles in cellular decision-making and providing the cells with a powerful capability to readily change their genomic expression in order to survive and reproduce successfully in ever-changing environments. For that reason, the field of epigenetics is considered a promising solution for cancer prediction and eradication. New discoveries in the epigenetic regulation of cells are gaining more and more attention in diagnosis. These epigenetic mechanisms have been shown to play important roles during cell development and equilibrium maintenance. Chatterjee and Zetters’2 study helped elucidate the shortcuts that cancer cells take by shifting these regulating routes for longer survival and proliferation over normal cells. Furthermore, several epigenetic changes occur prior to histopathological changes, constituting outstanding biomarkers for early cancer identification, diagnosis, and risk assessment.3 With a better knowledge of epigenetic mechanisms, one would be able to understand when, why, and how a tumour is produced. In addition, the specific reversion of these routes represents an excellent therapy solution in order to return the cancer cells to their normal state, without implying invasive chemotherapies and post-treatment side effects.
Currently, new diagnostic devices are being developed in order to decipher and keep track of these epigenetic mechanisms.4 These tools use highly advanced technologies that allow a simpler and more cost-effective diagnosis. The ultimate goal is to use the devices outside the laboratory for the continuous monitoring of the patient before and after treatment. Such devices may allow the simultaneous detection of multiple biomarkers belonging to a previously defined cancer epigenetic profile. In this context, nanotechnology is playing a decisive role by offering ultrasensitive devices with powerful and highly improved performances as compared to the current diagnostic techniques.5 The combination of nanotechnology and epigenetics is advancing towards a new generation of cancer diagnostic tools, where the primary objective is to provide low-cost, simple, and ultra-detailed analyses of the patients’ clinical conditions in record time. This early diagnosis will define a newer and hopeful starting point for cancer treatment, helping design specific protocols and targets for therapies specifically tailored for each individual.
This manuscript reviews the benefits provided by the study of epigenetic mechanisms and their relationship with cancer. We also present some emergent, brand new nanobiosensor technologies, aiming to provide an advanced diagnostic tool by analysing the epigenetic mechanisms altered in cancer. The new nanosensors accomplished the premises of user-friendly, reduced costs, and improved performance, priming them to spark a diagnostic revolution.
THE CANCER EPIGENETIC SIGNATURE
Different epigenetic mechanisms control the gene expression at different levels, constituting a complex, highly structured gene regulatory network. One of the most relevant epigenetic processes is DNA methylation. It consists of the addition of methyl groups to cytosine nucleotides (5-methylcytosine [5-mC]) in the DNA sequence to impede transcription, aiding cells to modulate the expression of the implicated genes at their convenience.5 In the same way, the cell can reorganise its chromatin structure with another epigenetic mark, the acetylation/deacetylation of its histones, affecting the transcription process. This epigenetic regulation of the transcription process has a strong influence at the post-transcription level, as in the case of the regulation of alternative splicing of messenger RNA (mRNA), which gives the cell the capability to alternatively edit a pre-mRNA to produce proteins with different functions, increasing diversity.6 On the other hand, cells can regulate the gene expression through micro-RNA. These small, single-stranded, non-coding RNA typically contain 19–23 nucleotides and play important roles in modulating several biological functions through their interaction with mRNA.7 In this way, cells will either translate or not a specific gene via the regulation of epigenetic control.
All these mechanisms are interconnected and play important roles in every process of life, including cell differentiation, metabolism, cell cycle, and signal transduction. Therefore, it is not surprising that their alteration disturbs the fate of cells and the consequences can be devastating, resulting in the origin of grave diseases, one of the most predominant of which is cancer. In fact, aberrant DNA methylation has been linked to the silencing of tumour suppressor genes, leading to the cancer development.8 Most cancers are associated with a switch in the splicing pattern of specific isoforms, providing them with a highly proliferative capacity and survival properties.9 Likewise, an aberrant expression of specific micro-RNA sequences has been directly linked to the generation of tumours.10 As a consequence, these epigenetic mechanisms are now being considered hallmarks of cancer. Current cancer biomarkers are based on overexpressed cancer proteins in blood; however, their number and clinical use are rather limited and require a large population of cancer patients with well-defined clinical staging and outcomes. Analysis of the epigenetic pathways may be more informative, specific, and accurate than the analysis of such protein biomarkers, by the determination of not only the cancer itself, but also the underlying mechanisms by which it is generated. Recent studies indicate that these regulation pathways participate in collaborative activities resulting in a common outcome.11 Understanding the dynamics of these networks can shed light on the mechanisms responsible for the development of many types of cancer. On top of this, it will contribute to a more efficient management of cancer patients, providing an early diagnosis, determining precise tumour staging, and monitoring of treatment.
The revolutionary epigenetic inheritance has changed how we understand and deal with cancer. The dynamic nature and potential reversibility of the epigenetic mechanisms mean that they are appealing therapeutic targets in cancer treatment. Applying therapies focussed on reversion of the altered processes to their normal state would abate the cancer progression in a less invasive and more efficient manner than standard chemotherapies. Currently, various compounds that can rearrange DNA methylation and histone acetylation patterns are being examined in clinical settings in combination with other drugs.12,13 Likewise, different approaches for mRNA and non-coding-RNA expression control are currently being assessed in order to potentially use them in more directed therapies.14-17 Therefore, a better knowledge of cell decision-making in cancer can be better exploited for the development and implementation of a more personalised medicine.
BIOSENSORS: THE DIAGNOSTIC SOLUTION
Advances in DNA/RNA detection techniques have recently hit the epigenetic field. Developments in real-time polymerase chain reaction (PCR) have allowed results to be obtained in a matter of a few minutes regarding the methylation status or mRNA isoform shift.18,19 DNA microarrays permit the interrogation of thousands of micro-RNA sequences simultaneously in one sample,20 while next-generation DNA sequencing technologies have expanded to genome-wide scale screening and have achieved a resolution to single base precision.21 However, these methods suffer technological limitations, such as slow turnaround times,22 relatively large input volume,23 and biases arising from sample contamination and PCR-induced artifacts.24,25 They usually require the consumption of expensive reagents in every assay and need trained personnel for their manipulation due to the complexity of the analytical protocols. Besides, all cancers involve more than one epigenetic mechanism, making the simultaneous detection of multiple epigenetic biomarkers essential.
Emerging trends in diagnostics have promoted the development of diagnostic tools with improved sensitivity and short operation times, such as biosensors. Biosensors can be designed to provide quantitative analytical information with elevated accuracy in a few minutes, using low sample volumes and minimum sample pretreatment. By definition, a biosensor is a self-contained analytical device that incorporates a biologically active material in intimate contact with an appropriate transduction element for the purpose of detecting, in a very selective way, the concentration or activity of chemical species in any type of sample (Figure 1).26
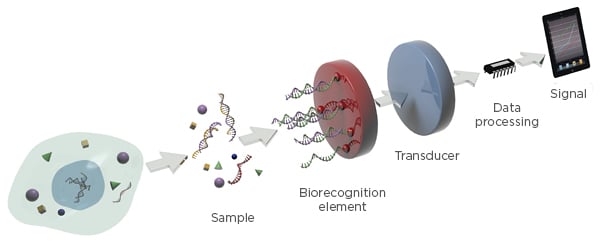
Figure 1: Schematic diagram of a biosensor device.
Adapted with permission from Carrascosa et al.31
Biosensors are threatening to radically alter our present concept of clinical analysis, beginning many years ago27 with the introduction of the glucose biosensor signifying a breakthrough in healthcare by the decentralisation and simplicity of analysis at home by one blood-drop.28 The principal aim of biosensors is to get away from the centralised laboratory and to provide analytical services closer to the patient: at the bedside, in the physician’s office, or by the patient at home.
Nowadays, there are some commercial biosensors for several applications, such as detection of clinical biomarkers or pathogens and toxic metabolites for environmental/food contamination and bio-threats.29 Biosensor devices can show extremely low detection limits, which permits the detection of biomarkers at their physiological concentrations, assisting in the diagnosis of cancer at very early stages. In the last decades, a particular interest has been focussed on the development of novel, label-free optical biosensors able to generate a signal directly by the interaction of the analyte of interest with the recognition element, without requiring additional interactions with other probes carrying a label that provides the signal. In general, label-free methods offer potential advantages in terms of simplicity and velocity of the bioanalysis, which may not require washing steps or additional reagents. These biosensors enable the real-time monitoring of the biomolecular interaction, speeding up detection and giving access to the kinetic parameters of the recognition process.30 Moreover, with the advent of micro and nanotechnology, more sophisticated label-free, optical biosensors combine extremely high-quality performances and ultrasensitive limits of detection with the ability to miniaturise and integrate different functional components such as microfluidics, electronics, etc. in a single platform. These characteristics allow for the fabrication of smaller, cheaper, and easy-to-use biosensor devices that can accelerate the real implementation of lab-on-a-chip devices in clinical practice.
Optical biosensors have shown exceptional capabilities for the detection of DNA/RNA sequences, at detection limits more than adequate for the physiological concentration of the target sequences in real samples.31 All optical biosensors developed so far for epigenetic biomarkers are characterised by the commitment to overcome the challenges encountered in their analysis (see Table 1 for a summary).
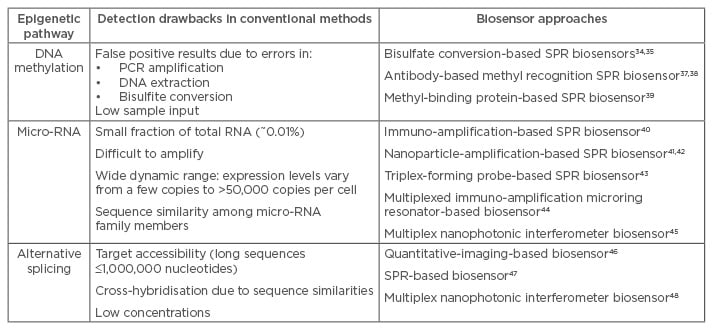
Table 1: Challenges in the biosensing of epigenetic mechanisms and different biosensor approaches.
PCR: polymerase chain reaction; SPR: surface plasmon resonance.
For instance, the modification of the DNA sequence by methyl-groups has been of interest in many optical biosensor applications.32 Some methodologies carried out analysis of DNA methylation through bisulfite conversion, employing a surface plasmon resonance (SPR) biosensor. This type of biosensor uses a gold layer as both a surface to immobilise specific probes and a transducer of the signals produced by the refractive index changes at the surface.33 Bisulfite converts non-methylated cytosine nucleotides into uracil nucleotides, allowing the identification of different methylated or non-methylated sequences by different amplification processes, such as PCR34 or molecular inversion probe amplification.35 However, bisulfite conversion does not differentiate between other epigenetic marks derived from enzymatic oxidisation of 5-mC, with important significance in the determination of the methylation status.36 Also, the possibility of bias introduced by errors in this pre-manipulation process has promoted the search for other alternatives. Specific recognition of methyl-sites (and derivatives) has been solved with specific antibodies37,38 or proteins39 being able to quantify the number of cytosines without the necessity of sample manipulation. In this way, new microfluidic designs have been proposed in order to provide smaller and more easy-to-handle equipment for DNA methylation analyses.38 The microfluidics incorporate different pre-treatment processes on the same microchip as the methylation analysis, such as fragmentation by restriction enzyme to obtain the fragment of interest, mixture with a biotinylated bulge-inducer probe to improve the methyl-group accessibility, and heat denaturation and cool down for hybridisation.
In the case of micro-RNA detection, due to their small size, short RNA regulators are difficult to amplify through conventional methods. In addition, they usually belong to a micro-RNA family with very similar sequences that can distort the analysis with false positive signals. The main objective towards micro-RNA detection relies on two premises: specificity and a high sensitivity to cover a wide range of concentrations. The ability to perform the experiments directly from an untreated biofluid without the need for purification steps, thereby risking sample input, is also important. Some SPR biosensors have been developed for the analysis of micro-RNA. Šípová et al.40 demonstrated the detection of specific micro-RNA in mouse liver tissues. To achieve the required sensitivity levels without PCR-amplification steps, researchers used a signal enhancer based on a specific antibody.40 Other signal enhancers have also been proposed, such as nanoparticles41,42 or specially designed probes, which promote a better target capture.43 Another approach based on microring resonator biosensors has been proposed.44 The miniature size of the microrings offers the possibility of an array of biosensors (32 microring resonators within a 6×6 mm footprint) for multiplexed measurements, which is very encouraging for their use in routine clinics. On the other hand, our team has designed a nanotechnology-based optical biosensor with multiplexing capabilities that has been able to cover the total analytical range from attomolar to nanomolar concentrations, skipping any further enhancement.45 The biosensor consists of optical waveguides, the nanometre dimensions of which change the properties of the guided light, generating ultra-sensitive interferometric signals able to detect attomolar concentration of micro-RNA directly in urine samples from bladder cancer patients. Its miniaturised size allows for multiplex arrays formats, incorporating 20 nanosensors within the same sensor chip (10 mm width, 31 mm length).
Few optical biosensors have been devised in order to use the alternative splicing regulation route for diagnostic purposes,6-8 probably due to the long RNA sequences and the similarity between mRNA isoforms that critically complicate the differentiation between the isoforms. Single mRNA-spliced variants have been identified and quantified in living cells by quantitative imaging.46 This advance may enhance the understanding of pharmacogenomics, genetic diagnosis, and gene therapies. Another methodology incorporated a fragmentation process in order to adapt the length of the isoforms to the biosensor convenience and standardise the detection procedure.47 The amplification-free methodology performed an accurate and efficient analysis of the alternative spliced isoforms from different genes and contexts in HeLa cells. In addition, the further implementation of the methodology in a multiplexed nanophotonic biosensor not only improved the sensitivity of the detection, but also would allow the simultaneous detection of an array of biomarkers with the same biosensor chip.48
CHALLENGES IN EPIGENETIC BIOSENSING
Despite the significant advances and the promising perspectives of these new emerging diagnostic optical biosensors for identifying cancer epigenetics, they still face several challenges. Epigenetics is a relatively young discipline and, as such, still more fundamental knowledge is needed for its translation into diagnostics as an accurate mark of cancer. Due to the immense network of different epigenetic routes, it is strictly indispensable to establish accurate and well-defined epigenetic panels to be correlated with concrete cancer profiles. These panels cannot give rise to possible misleading diagnostics and should have sufficient consistence to offer a robust diagnosis. Once this accurate definition takes place, many clinical trials have to be done in order to establish standard patterns between different individuals and set up threshold concentration that confirms the presence and spreading of a tumourigenic process. For cancer stratification, different stage profiles need to be predefined in order to know the stage and level of progression of the cancer.
On the other hand, the biosensor field is still far from meeting some of the key end-user needs. Most biosensor technologies should be focussed on matching or even improving their sensitivity levels to the conventional methodologies, without the need for sample premanipulation processes. Efforts are also focussed on the development of biosensors capable of multi-test detection and simultaneous monitoring. In terms of optical biosensor integration, new microfluidic approaches need to be addressed in order to overcome the specific requirements for each epigenetic mechanism and compartmentalisation for individualised analysis inside the same sensor platform. Also, the stability, reproducibility, and life cycle should be tested and guaranteed.
CONCLUSION
The benefits of optical biosensors for the routine analysis of epigenetic biomarkers is changing the concept of cancer diagnosis. The analysis of epigenetic regulation processes is a top requisite for the careful dissemination of tumour onset and the development of personalised treatments in order to reverse the affected mechanism in each cancer and patient. The possibility for rapid, very sensitive, and multiplex detection of different epigenetic mechanisms involved in tumour progression makes these devices an outstanding solution for cancer management. Several biosensor approaches have already been shown to fulfil the needs and overcome the challenges that conventional tools for epigenetic analysis are seeking nowadays. Further investigations are devoted to developing more robust, sophisticated, and easy-to-use devices. The combination of nanotechnology with epigenetic analysis constitutes a breakthrough in cancer theranostics, i.e. the use of one particular molecular diagnostic to guide therapeutic decisions, since it offers the possibility of not only providing a more in-depth vision of the tumourigenic process, but also paves the way for a more directed and effective route to design therapies aiming at eradicating cancer and increasing patient quality of life.








