Abstract
Since their introduction in 2001, tyrosine kinase inhibitors (TKIs) targeting BCR-ABL have become the standard therapy for chronic myeloid leukaemia (CML). While allogeneic hematopoietic stem cell transplant is a recognised curative treatment for CML, TKIs prevent progression to advanced phase in most patients, and spectacularly improve the disease burden (in deep molecular responders) and the overall survival of CML patients.
However, mutations in the BCR-ABL kinase domain affect a significant proportion of CML patients and have been associated with primary or secondary (refractory disease following an initial response) resistance to imatinib. Such resistance may emerge at any time during TKI therapy and are a major mechanism of treatment failure, in addition to BCR-ABL1-independent treatment resistance and treatment intolerance mechanisms. In the context of the above-described clinical settings, the management of CML patients remains challenging. The detection of mutations following imatinib resistance is therefore crucial to ensure appropriate second or third-line drug selection.
INTRODUCTION
Chronic myeloid leukaemia (CML) is a Philadelphia chromosome positive (Ph+) clonal bone marrow stem cell disorder classified into the group of myeloproliferative neoplasms, along with polycythaemia vera, essential thrombocythaemia, and primary myelofibrosis.1,2 CML originates from a single pluripotent haematopoietic stem cell, in which cells of the myeloid lineage undergo inappropriate clonal expansion caused by a molecular lesion.1,2
CML is characterised by the occurrence of the Philadelphia chromosome, which results from the fusion of the breakpoint cluster region (BCR) gene on chromosome 22 and the Abelson murine leukaemia (ABL1) gene on chromosome 9. This generates the BCR-ABL oncogene that encodes for a chimeric but active oncoprotein, the BCR-ABL tyrosine kinase; its deregulated activity is necessary and sufficient for malignant transformation.1,2 The disease typically progresses through three distinct phases: chronic phase, accelerated phase, and blast crisis, during which the leukaemic clone progressively loses its ability to differentiate.
Since their introduction in 2001, tyrosine kinase inhibitors (TKIs) targeting BCR-ABL have become the standard therapy for CML. While allogeneic hematopoietic stem cell transplant (Allo-HSCT) is a recognised curative treatment for CML, TKIs prevent progression to advanced phase in most patients, and spectacularly improve the disease burden (in deep molecular responders) and the overall survival of CML patients.3 At present, five TKIs are approved for the treatment of CML: imatinib (a first-generation TKI), nilotinib, dasatinib, bosutinib (second-generation TKIs), and ponatinib (a third-generation TKI). The first three compounds are approved for the treatment of newly-diagnosed patients who are treatment-naïve, while bosutinib and ponatinib are indicated in patients with resistant or intolerant CML.4-6
However, mutations in the BCR-ABL1 kinase domain (KD) affect a significant proportion of CML patients and have been associated with primary or acquired (refractory disease following an initial response) resistances to imatinib.6-9 Such resistance may emerge at any time during TKI therapy and are a major mechanism of treatment failure, in addition to BCR-ABL1-independent treatment resistances and treatment intolerance mechanisms.
In the context of the above-described clinical settings, the management of CML patients remains challenging. Indeed, while nilotinib and dasatinib are active against most imatinib-resistance mutations, other mutations also confer resistance (thus a poor response) to second-generation TKIs. Conversely, some imatinib-resistant mutations are insensitive to dasatinib and/or nilotinib.10-14 The detection of such mutations following imatinib resistance is therefore crucial to ensure appropriate second or third-line drug selection.15 Therefore, this article will review the available techniques to perform mutational analyses in CML patients, and how physicians can refine treatment selection pathways and rationales to select the appropriate therapy and tailor the management for each CML patient.
BCR-ABL MUTATIONS AND TREATMENT FAILURES
Mutations occur in cancerous cells where the genetic instability is high, leading to the accumulation of further abnormalities and evolution to advanced disease.16,17 In newly-diagnosed chronic phase (CP)-CML patients, 15–30% who start first-line TKI therapy will not reach an optimal response, and a BCR-ABL1 KD mutation will be detectable in 25–50% of patients with treatment failure.4,5,8,16,18-20 Furthermore, up to 80% of patients with blast phase (BP)-CML can carry mutations.21
Among BCR-ABL1 KD mutations, the T315I multiresistant mutant is found in 11-20% of cases.11,20,22-24 Small cell populations in which mutations occur may have a survival advantage during TKI therapy and emerge later as the dominant clone, speeding up the progression to AP-CML.16,17
Although resistance to therapy can occur at any time point, it has been established that the sequential use of TKIs as the CML disease progresses increases the probability of mutations.13,16,22,25 Relapsed patients usually display a greater genetic instability and have a higher likelihood to develop further mutations. As an example, a study showed that 83% of imatinib-resistant patients who relapsed while on a second or third-line TKI experienced an emergence of newly acquired BCR-ABL mutations.13 Moreover, the order of the TKI sequence may influence the emerging mutation type.25
SINGLE MUTATION AND COMPOUND MUTATIONS
Over 80 BCR-ABL KD single mutations that affect TKI sensitivity in CML have been identified with data collated from 27 studies, from patients resistant to first-generation TKI therapy.16,26 Compound mutations, defined as ≥2 mutations in the same BCR-ABL1 molecule (as opposed to polyclonal mutations, multiple BCR-ABL1 mutant clones) can confer high-level resistance to TKIs and are associated with suboptimal response and poorer outcomes, due to a very low TKI sensitivity.12,27-33 It has been suggested that sequential therapy with multiple TKIs may select for compound mutations that confer resistance to multiple TKIs.12,18,28
According to the type of mutation, corresponding TKI sensitivity can be observed, and in vitro potency of each TKI (IC50, corresponding to the concentration at which 50% of the BCR-ABL tyrosine kinase is inhibited) can be useful to predict which TKI could be more effective than others.34,35 As an example, Zabriskie et al.32 developed a heat map of IC50 values for single and compound mutants (Figure 1).
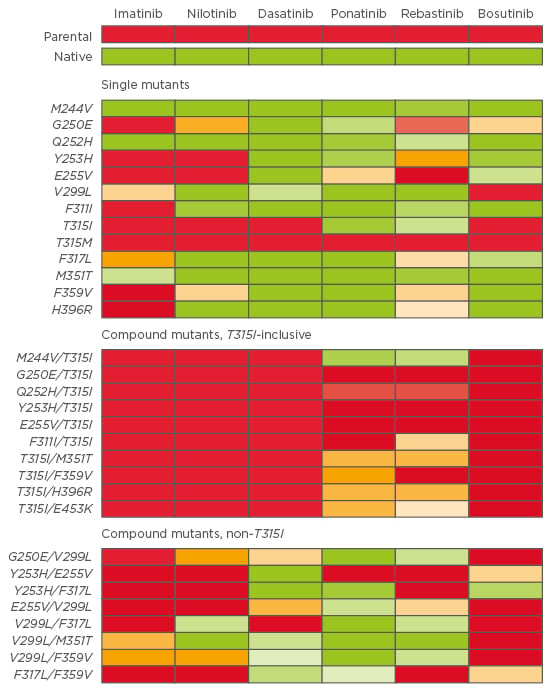
Figure 1: Heat map of TKI IC50 for single and compound mutants. A colour gradient from green (sensitive) to yellow (moderately resistant) to red (highly resistant) denotes the IC50 sensitivity to each TKI.32
TKI: tyrosine kinase inhibitor; IC50: concentration when inhibitor response is 50%.
TYPES OF MUTATIONAL ANALYSES
The presence of mutations is an important factor when making treatment decisions. Indeed, if an inappropriate TKI is chosen, there is a high-risk of subsequent treatment failure with clonal expansion of the resistant mutant, and a greater likelihood to select for a compound mutant: the initial mutated clone is not eradicated, thus has the possibility to acquire additional mutations.36 Several types of mutational assays have been developed to explore these mutational profiles in real-life clinical settings (absence/presence of mutations and mutation type) and their advantages and disadvantages are summarised in Table 1.9,37-40
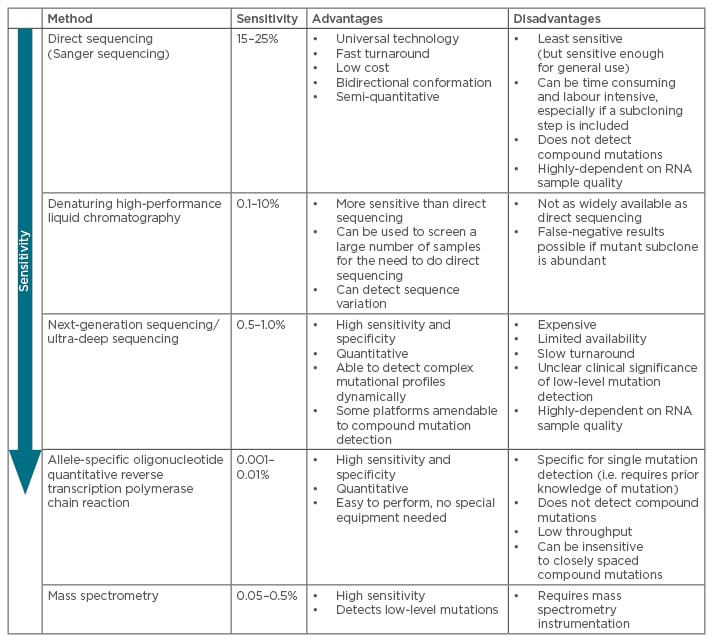
Table 1: Most common techniques for mutational analyses of BCR-ABL kinase domain mutations.9,36-41,58
The most common techniques for mutation screening of the entire KD are direct (Sanger) sequencing (SS) and ultra-deep sequencing (UDS) using next-generation sequencing (NGS). Assays for detection of given mutations include allele-specific oligonucleotide quantitative reverse transcription polymerase chain reaction (ASO-Rt-qPCR) and Sequenom mass spectrometry. Highly sensitive assays can be useful in predicting the best course of treatment for TKI-resistant patients and for monitoring resistant mutations in subsequent treatment settings.41
Sanger Sequencing
SS (direct sequencing) is the most common technique to detect BCR-ABL1 KD mutations associated with TKI resistance, as currently recommended by international guidelines and consensus panel.16,38,42 While being the least sensitive method available and associated to technical limitations, it has been deemed sufficient for general use by the haematological community, since it is widely available in laboratories worldwide.42 However, SS may not detect all mutations present, namely compound mutations and mutations present in less than 20% of cells (low-level mutations), below the detection limit. Mutations detectable by SS may just be the ‘tip of the iceberg’.38,40,41,43
Denaturing High-Performance Liquid Chromatography
Denaturing high-performance liquid chromatography is more sensitive (but not as widely available) than direct sequencing (0.1–10%), can detect sequence variation, and can be used to screen a large number of samples without the need to do direct sequencing.37 However, false-negative results can be generated if mutant subclone is abundant.
Next-Generation Sequencing and Ultra-Deep Sequencing
Deep-sequencing boasts a higher level of sensitivity (≥1%) to detect clinically relevant BCR-ABL emergent mutant clones that are not detected by SS, including compound mutations and the T35I mutation.43,44 Of note, NGS is the technology, while UDS (or direct sequencing) is the application of NGS for sensitive (deep) mutation screening of target genes (or gene panels). The increased sensitivity allows deep sequencing to qualitatively and quantitatively assess the clonal texture of the mutated BCR-ABL-positive subpopulations, giving the possibility to fully characterise the spectrum of mutants in a patient.40,45,46
In a cohort of 121 CP-CML patients presented at the 2015 American Society of Hematology (ASH) congress, we reported that NGS can reliably detect low-level KD mutations otherwise not detectable by SS. In particular, we found that NGS can detect low-level KD mutations in patients who achieve complete cytogenetic response (CCyR) but not major molecular response (MMR), thus allowing potential early clinical intervention.47 Finally, NGS could also detect the appearance of KD mutations as early as 3 months post TKI initiation in patients who failed to respond.
Soverini et al.48 recently reported the use of NGS to retrospectively screen a cohort of 60 imatinib-resistant patients (CML, n=45; Ph+ acute lymphoblastic leukaemia [ALL], n=15) who had failed second-line second-generation TKI therapy and acquired KD mutations (Group 1) compared to 25 imatinib-resistant patients (CML, n=21; Ph+ ALL, n=4) who had responded to second-line second-generation TKI therapy (Group 2).
The authors demonstrated that NGS was effective at detecting clinically-relevant mutations at the time of imatinib failure. In 43% of patients from Group 1, second-generation TKI-resistant mutations generating relapse were already detectable at low levels with NGS. When patients subsequently received a second-generation TKI therapy to which they were insensitive, mutations underwent clonal expansion in all cases. Conversely, no low-level mutation that was resistant to the second-generation TKI the patients subsequently received was detected in Group 2. This demonstrates that NGS at the time of imatinib failure could be efficient for more effective therapeutic tailoring and second-generation TKI therapy choice.
Allele-Specific Oligonucleotide Reverse Transcription Quantitative Polymerase Chain Reaction
ASO-Rt-qPCR boasts high sensitivity and specificity (the former at rates of 0.001–0.01%), and can be used for single mutation detection but not compound mutations. Its main drawbacks are a low throughput, restricted availability, and low sensitivity for closely-spaced compound mutations.20,49
Mass Spectrometry
Mass spectrometry is a more sensitive (detection limit of 0.05–0.5%) technique and was demonstrated to detect low-level mutations versus SS in patients following imatinib failure. Indeed, some mutations have been associated to resistances to nilotinib and/or dasatinib, and low-level mutations can influence failure-free survival (FFS), as demonstrated in a large study evaluating CP-CML patients treated with nilotinib or dasatinib after imatinib failure.36 In 220 CML patients with failure to imatinib, mutations that would influence therapeutic decisions and FFS were found in 71 patients with mass spectrometry compared to only 50 with SS (32% versus 23%; p=0.03; Figures 2 and 3).36
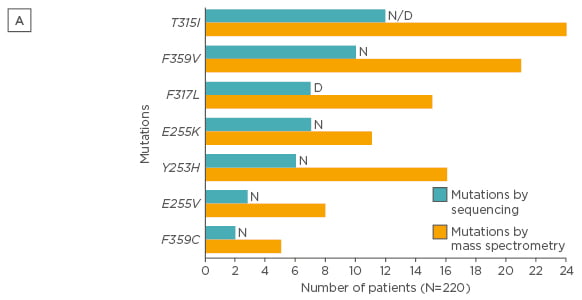
Figure 2A: Type of mutations detected by SS and mass spectrometry (only mutations that would influence therapeutic decisions after imatinib are presented).36
SS: direct (Sanger) sequencing; N: nilotinib; D: dasatinib.
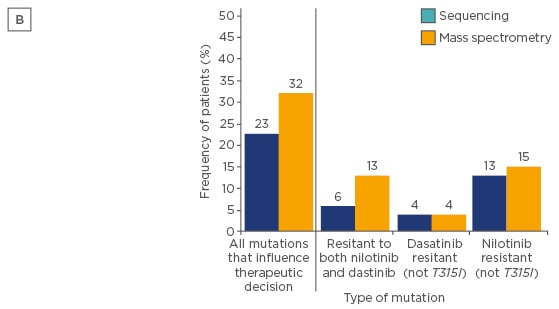
Figure 2B: Frequency of patients in whom one or more of their mutations detected at switchover would influence therapeutic decisions after treatment with imatinib failed.36
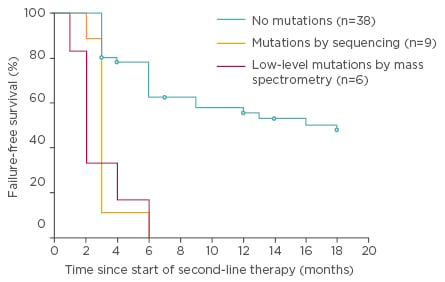
Figure 3: Failure-free survival by 18 months of nilotinib or dasatinib therapy for the 100 chronic phase patients according to mutation status at switchover.36
RECOMMENDATIONS IN PERFORMING MUTATIONAL ANALYSIS
European LeukemiaNet Recommendations
In a European LeukemiaNet (ELN) consensus meeting and article in 2011, experts stated that while mutations studies can help make treatment decisions in the context of patients presenting with AP/BP-CML at diagnosis, cytogenetic/haematologic relapse, or suboptimal response to first-line therapy, loss of MMR, there is currently no role for mutation analysis at diagnosis or in patients with adequate response to therapy.15,16,50
The 2013 ELN recommendations for the management of CML suggest mutational analysis should be performed with SS in case of treatment failure or progression to AP or BP-CML (Table 2).42 The 2013 ELN recommendations confirmed and replaced the term ‘suboptimal response’ with ‘warning’, so mutation analysis was recommended at diagnosis in patients presenting AP/BP, and in the case of failure or ‘warning’. Patients with a ‘warning’ response status require more careful and frequent monitoring, that is to say a molecular and a cytogenetic test within <3 months, along with a mutational analysis.
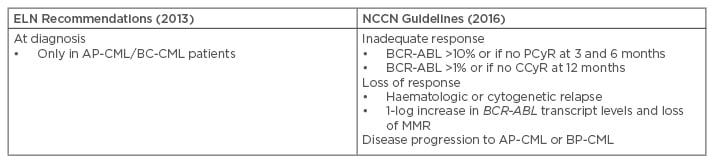
Table 2: Recommendations on when to perform mutational analysis.16,42,51
AP-CML: accelerated phase chronic myeloid leukaemia; CCyR: complete cytogenetic response; CP-CML: chronic phase chronic myeloid leukaemia; ELN: European LeukemiaNet; MMR: major molecular response; NCCN: National Comprehensive Cancer Network; PCyR: partial cytogenetic response; BP-CML: blast phase chronic myeloid leukaemia.
National Comprehensive Cancer Network Guidelines
The 2016 National Comprehensive Cancer Network (NCCN) Guidelines ascertain that routine monitoring of BCR-ABL transcripts, in conjunction with cytogenetic evaluation, provides important information about long-term disease control in patients with CML.51 These guidelines state that mutational analysis should be conducted in patients who fail to achieve first-line TKI treatment targets, who lose response, or who progress to AP-CML or BP-CML (Table 2).51 Of note, NCCN guidelines do not recommend a specific technique, while the ELN guidelines recommend SS.42,51
Some authors have suggested the importance of conducting mutational analysis in patients with resistances either while maintaining the patient on TKI therapy or just before stopping/switching TKI therapy. Indeed, should TKI therapy be discontinued, the results of the mutational analysis (and detection of underlying mutant copies) could be biased by the proliferation of non-mutated BCR-ABL1 cells without kinase inhibition.18 One of the suggested cut-offs for mutation analysis in the literature, including NCCN guidelines,38,51 is a 5 to 10-fold increase in BCR-ABL1 transcript levels and loss of MMR, which can be put in perspective with the findings of a study conducted in 150 patients receiving imatinib as first-line therapy.52 The investigators observed that a 2.6-fold rise in BCR-ABL1 transcript levels was associated to the emergence of BCR-ABL mutations, Moreover, transcript rise cut-offs of 5-fold or greater had poor diagnostic sensitivity and no significant association with mutations, which could suggest that such thresholds are insensitive and not universally applicable.
THE IMPORTANCE OF MUTATIONAL ANALYSIS FOR TREATMENT OUTCOMES
Despite being recommended in current treatment guidelines, mutational analysis is not always performed in patients with suspected TKI resistance and a repeated screening is rarely done in patients proven to be previously negative for BCR-ABL1 KD mutations. Physicians do not always test for mutations when appropriate, or for economic reasons, and many do not appreciate the role of mutation analysis in the overall management of CML.18 In a prospective, non-interventional, cross-sectional study conducted in December 2010 through an online survey of 507 physicians treating patients with CML,53 nearly half of physicians did not test for KD mutations in patients not achieving a MMR 2 years after the initiation of TKI therapy. Also, 9% indicated that they were unfamiliar with or had never ordered a test for KD mutations.
This could be explained by the fact that both ELN and NCCN recommendations/guidelines provide only general recommendations to evaluate patients with resistance to TKI therapy. While patients being resistant to first-line therapies clearly require a closer evaluation of their mutation profile, ELN and NCCN guidelines do not specify the most appropriate testing technique according to the clinical context. This lack of precise, harmonised guidance could partially explain why a substantial proportion of physicians do not use mutational analysis to guide their decisions.
CHOOSING THE RIGHT TYROSINE KINASE INHIBITOR FOR THE DETECTED MUTATION
As stated above, the type of mutation present can help determine appropriate subsequent therapy. The results of mutational analysis are one of many factors (e.g. efficacy, safety, patient comorbidities, cost) in making treatment decisions.18 For patients with TKI-resistant CML, potential treatment options include an alternative TKI, protein synthesis inhibitors (omacetaxine, not approved in Europe) or ASCT.42 Following first-line failure, the NCCN have elaborated treatment recommendations based on BCR-ABL1 mutations (Table 3).51 The T315I mutant has shown resistance to all currently available TKIs, with the exception of ponatinib.16,32,42 Ponatinib is a third-generation TKI29 that has demonstrated clinical activity in the PACE Phase II trial, conducted on heavily pre-treated CML patients with or without KD mutation, and including the T315I mutant.54,55 Current data seem to indicate that secondary resistance to ponatinib is scarce, only occurring in patients with advanced CML.32,55
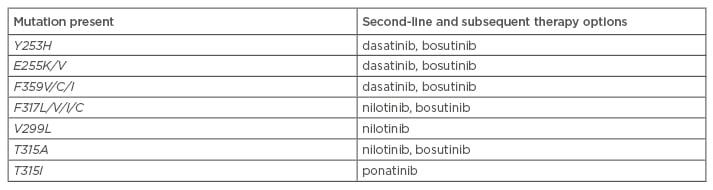
Table 3: National Comprehensive Cancer Network treatment recommendations based on BCR-ABL mutations.51
Ponatinib could also be of importance in patients with multiple mutations (and without the T315I mutation) following TKI resistance, as compared with nilotinib or dasatinib as second-line treatment modalities, which generate inferior responses. In a subset analysis conducted on 267 heavily pre-treated CP-CML patients from the PACE Phase II trial, NGS was performed to define baseline BCR-ABL1 mutation status.43 SS was also conducted to identify clonally dominant mutants that may have developed on ponatinib therapy (30.1-month median follow-up). Robust and durable cytogenetic and molecular responses were observed regardless of the technique (NGS or SS) and irrespectively of baseline mutation status. No single or compound mutation was identified as consistently conferring resistance to ponatinib in this cohort, which included patients with low-level T315I and compound mutations.
These results indicate that ponatinib could be effective in CP-CML irrespective of baseline mutation status, including the T35I variant and compound mutations. In such clinical settings, NGS may have a role in patient selection, namely those with low-level T315I and susceptible to benefit from salvage ponatinib therapy following second-generation TKI failure.43 To date, ponatinib is the most potent TKI and clinical data indicates rapid, deep, and durable clinical and molecular responses. However, considerable cardiovascular adverse events that could be dose-dependent should be taken into account to maximise the benefit-to-risk ratio.56,57
CONCLUSIONS
BCR-ABL1 KD mutations may emerge at any time during TKI therapy and confer treatment resistance, thus warranting the need for proper detection and selection for appropriate subsequent therapy. Mutational analysis is not always performed in patients with suspected TKI resistance, but should be considered a standard part of monitoring CML patients treated with TKIs. Moreover, mutational analysis should also be considered a standard adjunct evaluation tool in clinical research, especially in the context of the advent of promising new agents, such as ponatinib, to address multiresistance.








