BACKGROUND AND AIM
Occupational exposure to vapours, gases, dusts, and fumes (VGDF) has been associated with lower lung function1 and a higher risk of chronic obstructive pulmonary disease (COPD).2 The proportion of COPD attributable to occupational exposures has been estimated at 14%.3 A proposed mechanism underlying the effect of VGDF on COPD is through oxidative stress. A reduced antioxidant capacity has been found in COPD patients4 and higher levels of oxidative stress biomarkers have been associated with exposure to VGDF.5 The effect of VGDF on the lung might be apparent (or might be substantial) only in the presence of genetic variation (e.g., single nucleotide polymorphisms [SNP]) in antioxidant genes, indicating a gene–environment interaction. The study aimed to identify interactions between variants in well-known antioxidant genes and VGDF exposure on COPD risk.
METHODS
To assess antioxidant gene–VGDF interactions, the authors included 199,607 working individuals from the large UK Biobank (UKB; aged 39–71; 49% male; 44% never-smoker). Of these, 16,389 individuals had COPD spirometrically defined as forced expiratory volume in 1 second (FEV1)/forced vital capacity (FVC)< lower limit of normal (GLI-2012) and 34% were exposed to VGDF estimated using the airborne chemical (ACE) job exposure matrix.6 All independent SNP within 15 antioxidant genes selected based on a recent review were analysed.7 Gene–VGDF interactions were invested using a case-only approach using BOLT-LMM adjusted for sex, age, height, smoking status, genotyping batch, and centre, and accounting for population stratification/relatedness. Analyses were stratified according to smoking status. For significant interacting SNP, the authors assessed effects on gene-expression and DNA methylation levels using the online tool PhenoScanner.8
RESULTS
Overall, subjects exposed to VGDF had a 7% increased risk of COPD (Table 1a). However, this increase in risk was only seen in ever-smokers and was significantly different to never-smokers (interaction p=8.79×10-5). In the SNP–VGDF interaction analyses, the authors identified nine nominally significant interactions (p<0.05) for the antioxidant genes CAT (two SNP), GC (two SNP), GSTP1, GSTT1, NOS1, SOD1, and SOD2. Except for a SNP in GC, none of the SNP had a marginal effect on COPD and would therefore not be found if ignoring the effect modification by VGDF. Of the nine SNP, only two rare SNP with minor allele frequencies <3% in GSTP1(rs8191445) and NOS1 (rs145671209) survived correction for multiple testing (false discovery rate <5%). Subjects exposed to VGDF and carrying a copy of the minor allele of the GSTP1 and NOS1 SNP had a respective 19% and 30% higher risk of COPD compared to subject not exposed to VGDF and not carrying a copy of the minor allele (Table 1b). Although the interactions were only significant in never-smokers, the three-way SNP–VGDF–smoking interactions were not significant (p=0.338 for GSTP1 and p=0.460 for NOS1). The GSTP1 SNP was associated with expression of two nearby genes (DOC2GP/NDUFS8) in whole blood. No association was found with DNA methylation for either SNP.
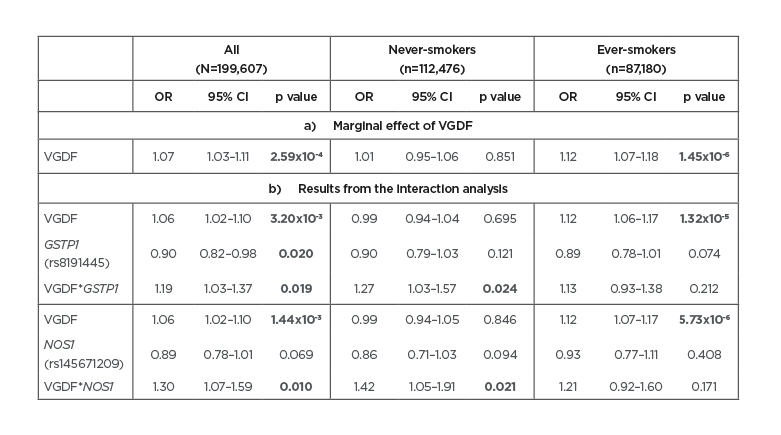
Table 1: Results of the association analysis of A) VGDF with COPD (marginal effect of VGDF); and B) of the SNP-VGDF interaction analyses for both the GSTP1 and the NOS1 SNP, stratified by smoking.
Significant results are presented in bold.
CI: confidence interval; COPD: chronic obstructive pulmonary disease; OR: odds ratio; SNP: single nucleotide polymorphism; VGDF: vapours, gases, dusts, and fumes.
CONCLUSION
These results suggest that VGDF exposure is associated with a higher risk of COPD, but that this risk may be confined to smokers. Those with rare variations in the antioxidant genes GSTP1 and NOS1 may be more susceptible to the effects of VGDF on COPD, and this effect might be more apparent in never-smokers. Further work is needed to replicate these results in independent samples and to investigate the possibility of three-way SNP–VGDF–smoking interactions.