Abstract
Objectives: To evaluate changes in microbial composition and the evolution of gastrointestinal tract (GIT) symptoms in systemic sclerosis (SSc).
Methods: Adult SSc patients provided stool specimens every 3 months over the course of 1 year. Participants completed the University of California, Los Angeles (UCLA) GIT 2.0 questionnaire to assess GIT symptom severity at each stool collection. The microbiota from these samples were determined by Illumina HiSeq 2500 16S ribosomal RNA sequencing (Illumina, Inc., San Diego, California, USA). Mixed effect models evaluated changes in GIT symptoms and microbial composition over time.
Results: Among 19 patients with SSc (female; 89.5%; median age: 51.3 years), the median disease duration was 7 years and the baseline total GIT 2.0 score was 0.7 (standard deviation: 0.6). The majority of participants (63%) provided at least four stool samples over the course of the 12-month study. Patients with longer disease durations had increased GIT symptoms over the course of the study. There was no difference in the course of GIT symptoms over time between patients with limited versus diffuse cutaneous disease. The relative abundances of specific genera did not change over time within individual subjects. After controlling for age, sex, ethnicity, disease duration, and SSc subtype (i.e., limited versus diffuse), low abundance of Bacteroides was associated with increased GIT symptoms over time.
Conclusion: This study is the first to have longitudinally characterised the lower GIT microbiome in SSc patients and demonstrated relative stability of genera abundance over the course of 1 year. The findings provide additional evidence that specific genera are associated with SSc-GIT symptoms and warrant further evaluation in larger SSc studies.
INTRODUCTION
Understanding the interplay between the immune system and gastrointestinal tract (GIT) microbiota is an evolving area of research.1,2 Emerging evidence suggests that GIT microbiota affects immune function in autoimmune diseases such as inflammatory bowel disease3 and rheumatoid arthritis.4
The authors’ groups and several others have discovered alterations in the GIT microbiota (i.e., dysbiosis) within distinct systemic sclerosis (SSc) cohorts.5-11 For instance, the SSc disease state is associated with increases in pathobiont organisms (e.g., Fusobacterium5,6 and Akkermansia5,6), and decreases in commensal organisms (e.g., Faecalibacterium,5-7Bacteroides,6 and Clostridium5,6 genera). The precise immunological implications of these changes in SSc are unclear; however, studies have demonstrated that commensal organisms, such as Bacteroides fragilis, induce and maintain regulatory T cells.12B. fragilis has been found to modulate immune cell responses through the secretion of outer membrane vesicles.13
In the authors’ prior work, they found that a decreased abundance of B. fragilis was associated with increased SSc-GIT symptoms.5 They also discovered that increased abundance of the pathobiont Fusobacterium was associated with increased GIT symptoms.5 Symptoms of lower SSc-GIT dysfunction include constipation, abdominal pain, diarrhoea, faecal incontinence, and/or weight loss,14 which all have the potential to contribute to psychosocial dysfunction and poor quality of life.15,16
However, the aforementioned studies were cross-sectional; therefore, it is unclear whether the relationships observed between specific genera and GIT symptoms are causational and/or persist with time. To address these questions, the present study sought to longitudinally characterise the GIT microbiome in SSc patients over a 12-month period while simultaneously evaluating GIT symptoms using a valid patient-reported outcome.
The objectives of this study, not previously explored in the SSc literature, were to: 1) evaluate changes in GIT microbiota of SSc patients over time; 2) assess changes in GIT symptoms in SSc patients over time; and 3) evaluate the hypothesis that specific microbial genera are associated with the characteristic symptoms of lower GIT dysfunction in SSc over time.
METHODS
Study Participants
Patient participants were consecutively enrolled from University of California, Los Angeles (UCLA), Los Angeles, California, USA, rheumatology clinics. Eligible participants included adult patients (â¥18 years) with SSc.17 Exclusion criteria included inflammatory bowel disease and the inability to withstand from taking an antibiotic and a probiotic for at least 3 weeks prior to the stool collection. Patients were allowed to continue their proton-pump inhibitor use because this medication appears to exert negligible effects on colonic microbiota,18 and abstaining from proton-pump inhibitor use in SSc patients can cause undue harm. As this was a proof of concept study to understand the dynamics of microbial composition in SSc patients over 1 year, there was no target sample size. The UCLA Institutional Review Board approved the study protocol and written informed consent was obtained from each participant.
Specimen Procurement and Processing
Participants collected stool specimens every 3 months for 12 months, using the authors’ previously published collection method,19 and the samples were immediately frozen. Frozen specimens were subsequently transferred on ice to the participant’s study centre. The study coordinator confirmed that specimens were still frozen at the time of arrival and immediately stored them at -80°C. Please see the authors’ previous publication5 for complete details of specimen procurement and processing.
16S Ribosomal RNA Gene Sequencing and Microbial Composition Analysis
The microbiota from the stool specimens were profiled by multiplex sequencing for bacterial 16S ribosomal RNA genes using an Illumina HiSeq 2500 (Illumina, Inc., San Diego, California, USA). All samples were analysed simultaneously to avoid any batch effects. The exact details of this approach have been outlined in our prior publication.5
To compare the microbial communities of SSc samples within each subject, measured every 3 months, alpha and beta diversity analysis were performed in QIIMEâ¢. Alpha diversity was assessed by the Chao1 index (metric of species richness) and Shannon index (metric of both richness and evenness). Repeated measures analysis of variance (ANOVA) was used to determine the significance of changes in alpha diversity within each subject. Beta diversity represents differences in microbial composition between samples and was measured using a robust Aitchison distance metric calculated using the DEICODE plugin in QIIME. Principal coordinates analysis was performed to visualise the resulting distance matrix. Significance was determined by permutational multivariate analysis of variance using the Adonis function in the R vegan package.
Assessment of Gastronintestinal Tract Symptoms
On the day of their stool collection, the SSc participants completed the UCLA GIT 2.0 questionnaire, a valid measure of GIT symptom severity in patients with SSc.20 The questionnaire consists of seven domains and has been validated in several independent SSc cohorts.
Statistical and Bioinformatics Analyses
Analyses were performed using R version 3.1.2. Mean and standard deviation (SD) were used to describe continuous parametric data and median and interquartile ranges were used to describe continuous nonparametric data. All tests were 2-sided with a 0.05 alpha level. The false discovery rate (FDR) correction method of Benjamini and Hochberg21 was used and a significant association was defined at the FDR q value â¤0.1.
To determine which baseline characteristics were associated with the course of GIT symptoms, each of the following variables were entered one at a time into the mixed models for the longitudinal assessment of GIT symptoms: age, sex, race, ethnicity, diffuse versus limited cutaneous disease, disease duration (years), Scl-70 positive versus negative, anticentromere antibody positive versus negative, presence or absence of interstitial lung disease (ILD) based on high-resolution CT, current prednisone use, and current other immunosuppression use. Among these variables, only disease duration and the presence of limited versus diffuse cutaneous disease were associated with the outcome. Subsequently, both of these variables (i.e., disease duration and the presence of limited versus diffuse cutaneous disease) were simultaneously included in the mixed effects model analysis for the total GIT symptom score and each individual domain score.
Mixed effects models were generated for each individual genus with each patient as a random effect to determine whether the relative abundance of specific genera changed over time. As some genera were present/absent in different patients, we also generated mixed effects models for longitudinal dichotomous data via GLIMMIX, where the outcome was the presence/absence of a particular genus.
Because neither the abundance, nor the presence/absence of any genera significantly changed over time, the average genus abundance level across all stool collections for each patient was calculated. The average abundances were included as a covariate into a rank regression analysis with the GIT score as the outcome. The other covariates in the model were disease duration and limited versus cutaneous disease. The clinical variables were first entered into the model before entering the genus abundances. This analysis was performed for each individual genus, and the results were adjusted for multiple hypothesis testing.
RESULTS
Participant Characteristics
A total of 19 patients with SSc (female: 89.5%) provided baseline stool samples and completed the UCLA GIT 2.0 questionnaire. The median age was 51.3 years and the median disease duration was 7.0 years (Table 1).
The majority of patients had limited cutaneous disease (68.4%) and had ILD based on high-resolution CT of the chest (73.7%). Few patients consumed immunosuppressant medications during the study (Table 1).
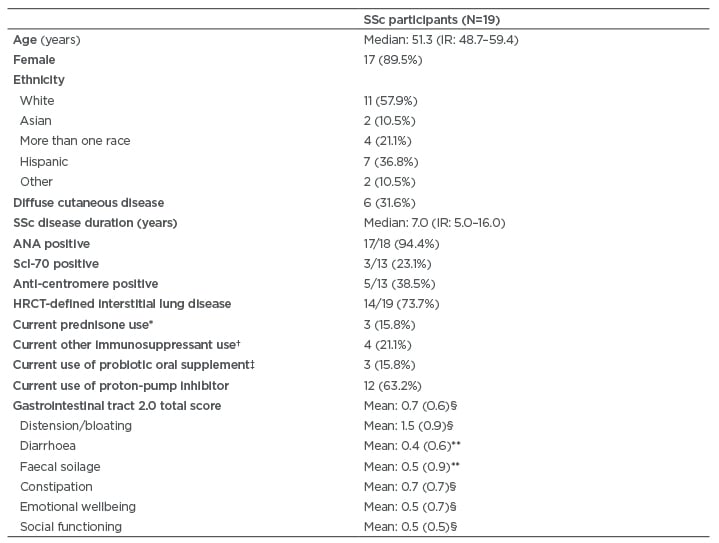
Table 1: Demographic and disease-related characteristics of systemic sclerosis participants.
Values are n (%), except where otherwise noted.
* Dosages of prednisone was ≤10 mg daily.
† Immunosuppressant medications used included mycophenolate (n=2) and azathioprine (n=2).
‡ Probiotics used included Culturelle® (Amerifit, Inc., Cromwell, Connecticut, USA) (n=1), Florify® (Melaleuca, Idaho Falls, Idaho, USA) (n=1), and Align® (Procter & Gamble, Cincinnati, Ohio, USA) (n=1). Probiotics were not consumed for at least 3 weeks prior to the fecal sample collection.
§ Score indicates moderate symptom severity.12
** Score indicates mild symptom severity.12
ANA: antinuclear antibodies; HCRT: high-resolution computed tomography; IR: interquartile range; Scl-70: topoisomerase 1; SSc: systemic sclerosis.
None of the patients used tobacco products. Alcohol use was also reported infrequently; five patients reported that they had consumed alcohol within the month prior to the baseline stool collection. For the remaining stool collections, six reported alcohol consumption at Month 3, and three reported alcohol consumption at Months 6, 9, and 12, respectively. Alcohol consumption did not exceed three servings per week in any patient. Patients reported no changes in dietary patterns or restrictions during the study. Patients also reported no changes in their household (persons and pets living with them, or location of their home).
Longitudinal Stool Specimen Collection
The majority of participants (63.2%) provided at least four stool samples over the course of the 12-month study (provided five samples [n=6]; provided 4 samples [n=6]; provided three samples [n=2]; provided two samples [n=4]; provided one sample [n=1]). The patient who only provided one sample was not included in the longitudinal analysis. Two patients died during the study (both male) from respiratory failure caused by ILD. For the remaining participants, the reason for incomplete stool collection was forgetting to bring their sample.
Antibiotic Use During The Study
Six patients (31.6%) had taken antibiotics in the 3 months preceding the baseline stool sample collection; the mean time between cessation of antibiotics and stool collection was 6.5 weeks (range: 4.0-12 weeks). For the remaining sample collection time points, all patients stopped antibiotics at least 4 weeks prior to the sample collection with the exception of one patient who was taking ciprofloxacin at the time of the sample collection (3-month sample collection: one stopped 4 weeks prior, one stopped 6 weeks prior, one was on ciprofloxacin as mentioned above; 6-month sample collection: one stopped 8 weeks prior; 9-month sample collection: two stopped 4 weeks prior, one stopped 5 weeks prior; 12-month sample collection: one stopped 4 weeks prior). The indications for antibiotic use were predominantly infections (specifically digital ulcer infections), except for the one patient who received ciprofloxacin during the 3-month collection for the treatment of small intestine bacterial overgrowth. Only three patients reported consuming a commercial probiotic at baseline (Table 1), and all discontinued the probiotic at least 3 weeks prior to each stool collection.
Evolution of Gastrointestinal Tract Symptoms Over 1 Year
At baseline, the mean UCLA GIT 2.0 scores indicated moderate symptom severity15 for the total score, as well as for the following specific domains: distension/bloating, social functioning, emotional wellbeing, and constipation (Table 1). The mean UCLA GIT 2.0 scores for faecal soilage and diarrhoea indicated mild symptom severity15 (Table 1).
In the mixed effects model analysis, total UCLA GIT 2.0 score did not change significantly over the course of the 12-month study (p value for time trend: 0.555). Individual UCLA GIT 2.0 domain scores also did not change significantly over the course of the 12-month study, including the scores for constipation (p value for time trend: 0.617), distension/bloating (p value for time trend: 0.726), diarrhoea (p value for time trend: 0.115), social functioning (p value for time trend: 0.333), emotional wellbeing (p value for time trend: 0.898), and faecal soilage (p value for time trend: 0.345).
Disease Duration Affects the Course of Gastrointestinal Tract Symptoms
In univariable mixed effects model analysis, the only baseline characteristics associated with the course of the total GIT score and the GIT scores for the individual domains were disease duration and SSc subtype (limited versus diffuse disease). When disease duration and SSc subtype were included as covariates in the models, only disease duration remained associated with the outcome (Table 2). Specifically, patients with longer disease durations had increased symptoms over time for the following: total GIT score, bloating, faecal soilage, diarrhoea (trend), social functioning, emotional wellbeing, and constipation.
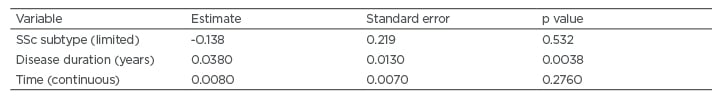
Table 2: Baseline characteristics associated with the course of total systemic sclerosis-gastrointestinal tract symptoms based on mixed effects model analysis.
SSc: systemic sclerosis.
Longitudinal Assessement of the Gastrointestinal Tract Microbiome
Microbial composition remained stable within subjects over the course of 1 year. Specifically, there was no significant difference in alpha diversity over the course of the study as measured by Chao1 index (p value: 0.78) and by the Shannon index (p value: 0.76). There was also no significant association between UCLA GIT 2.0 score longitudinally with the Chao1 and Shannon indices (p value: 0.44 and 0.27, respectively). The beta diversity analysis also showed no significant within-subject changes over the course of the study (p value: 0.68) and no significant association with UCLA GIT 2.0 score longitudinally.
Moreover, mixed effects models generated for each individual taxon at the genus level demonstrated that the relative abudance of each genus did not change significantly over time (all q values: >0.1). Moreover, if a specific genus was present or absent in a given subject at baseline, it remained present or absent at all of the subsequent analysis time points throughout the year-long study. Thus, this study found no change in individual taxonomic abundances at the genus level within each subject over 1 year.
Specific Genera are Associated with Gastrointestinal Tract Symptoms Over Time
Because GIT score and the abundance of specific genera did not change significantly with time, GIT scores and genus level abundances were averaged across all time points for every subject and entered into a rank regression analysis. The results of this analysis revealed that specific genera were associated with the course of GIT symptoms over 12-months, even after controlling for age, sex, ethnicity, SSc type (i.e., limited versus diffuse), and SSc disease duration. For example, increased abundance of Bacteroides, Prevotella, and specific genera from the Clostridiales order were associated with improvement in the course of total GIT symptoms over 1 year (Table 3).
DISCUSSION
This is the first study to longitudinally characterise the GIT microbiota of patients with SSc. The findings suggest relative stability of the GIT microbiome in SSc patients over the course of 1 year, with no appreciable changes in alpha and beta diversity or the relative abundance or presence/absence of specific genera. The study also demonstrated that self-reported GIT symptoms also do not change significantly over the course of 1 year in patients with SSc. However, consistent with prior studies,5,6 specific genera were associated with GIT symptoms.
The observation that GIT symptoms did not change significantly over the course of 1 year within subjects was initially suprising because many patients with SSc experience day-to-day variations in the severity of their GIT symptoms. However, since the GIT 2.020 asks patients to recall symptoms over the course of 1 week, it is plausible that these day-to-day variations were not captured. Since few studies have evaluated longitudinal progression of GIT symptoms, it is also possible that progression occurs at a relatively slow rate in patients with SSc. A study of longer duration (>1 year) may be needed to accurately estimate the rate of progression of GIT symptoms in patients with SSc.
The only variable found to affect the progression of SSc-GIT symptoms in this cohort was disease duration. Specifically, patients with longer disease duration had increased GIT symptoms over time. SSc subtype was not associated with the course of GIT symptoms when disease duration was considered. In line with these findings, Lock et al.22 reported that the prevalence of lower GIT symptoms were similar between patients with diffuse and limited cutaneous disease. The results of this study suggest that GIT symptoms should be carefully monitored in all patients, regardless of their SSc cutaneous subtype; however, patients with longer disease duration may require more vigilant monitoring.
Another unexpected finding of the present study was that the presence/absence of specific genera, as well as the relative abundances of specific genera, did not change significantly over the course of the study. These findings may be because patients consumed a relatively stable Western diet throughout the study period. While seasonal microbiota variations have been observed in some populations, such as the Hadza in Tanzania,23 such variations would not be expected among most individuals living in the USA. None of the patients reported significant changes in dietary patterns (e.g., adopting a gluten-free diet or plant-based diet for instance) that have been found in prior to studies to influence GIT microbiota.24,25
Consistent with prior studies using colonic lavage specimens5 and faecal specimens,6 certain genera were associated with the course of the GIT symptoms. Specifically, increased abundance of Bacteroides was associated with decreased total GIT score over time. This finding is consistent with a prior study, which demonstrated that patients with higher B. fragilis in both the cecum and sigmoid regions of the colon had decreased bloating/distension, diarrhoea, and total GIT symptoms.5 Considered a commensal genus by several host inflammatory and physiologic endpoints in animal models, Bacteroides was significantly lower in SSc patients from two geographically distinct cohorts (e.g., Oslo Univeristy Hospital [OUH], Oslo, Norway; and UCLA, Los Angeles, California, USA) compared to healthy controls.6 The fold change scores for both the UCLA-SSc and OUH-SSc cohorts relative to controls was nearly five, signifying a substantial shift in this genus in SSc.6 Low relative abundance of this genera is associated with increased disease activity in other autoimmune diseases, including Crohn’s disease.26 The present findings suggest that through increasing the abundance of Bacteroides, patients with SSc may potentially experience an improvement in GIT symptoms, and future interventional studies are needed to test this hypothesis.
In addition to Bacteroides, specific genera from the Clostridiales order (e.g., Finegoldia, Peptoniphilus, and Pseudomonas) were associated with decreased distension/bloating (see Table 3 for further details). In prior studies,5,6 other genera from the Clostridiales order (namely, Clostridium were associated with decreased total GIT symptom severity score, as well as GIT scores for constipation and the bloating/distension.5,6 In addition, Clostridiaceae was decreased among SSc patients with dysbiosis in a Swedish cohort study compared with eubiotic patients.7 Future therapeutic efforts may also consider increasing the abudance and/or activity of genera from the Clostridales order.
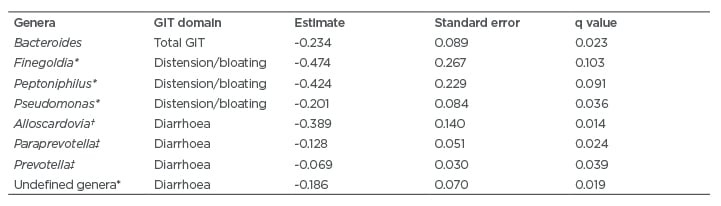
Table 3: Summary of microbial genera associated with decreased gastrointestinal tract symptoms over 1 year in patients with systemic sclerosis.
* Genera from the Clostridiales order.
† Genera from the Bifidobacteriales order.
‡ Genera from the Bacteroidales order.
GIT: gastrointestinal tract.
The findings of the present study should be considered within the context of certain limitations. First, the sample size is small, and the study may not be adequately powered to detect significant changes in symptoms/microbial composition over time. However, despite the small sample size, the significant associations between genera and symptoms were consistent with associations reported in prior studies,5,6 suggesting that the present findings are unlikely to be caused by chance alone. Second, to evaluate changes in specific genera over time, numerous inference tests were performed, which may have inflated the Type 1 error rate. To mitigate the risk of Type 1 error, the study used a relatively conservative FDR to correct for multiple hypothesis tesing. In addition, this study did not include a validation cohort.
Furthermore, this study did not include an objective assessment of GIT transit. Andreasson et al.7 found that the degree of dysbiosis correlated with the degree of dysmotility (based on cineradiography of oesophageal function). No studies have evaluated the effects of lower GIT dysmotility on microbiota in SSc. It is unclear whether dysmotility or the GIT pathology of SSc itself drives changes in microbial composition, or if it is the changes in microbial composition influencing GIT motility in SSc. To answer this question, longitudinal studies are needed, which will evaluate changes in microbial composition in conjunction with changes in motility over time.
In addition, it may be prudent to systematically assess dietary intake patterns in future SSc-GIT microbiome studies. In a relatively large study (N=1135), Zhernakova et al.27 identified 60 dietary factors affecting variations observed in the inter-individual distance of microbial composition. Assessing dietary intake is challenging because of recall bias; however, dietary modifications represent an important and low-risk means of altering GIT microbial composition and metabolic output.28,29 For instance, adoption of a diet low in fermentable oligosaccharides, disaccharides, monosaccharides, and polyols (FODMAP) led to signficant alterations in the microbiome metabolome of patients with irritable bowel syndrome.30
The present study also has important strengths. By studying microbial composition at multiple time points, the likelihood that the observed associations are caused by chance alone has been minimised. In addition, by performing all of the sequencing analyses simultaneously, the possibility of a batch effect has been eliminated. The authors also took caution to ensure that all patients withheld medications, such as antibiotics and probiotics, at least 3 weeks prior to the stool collection, by verifying the medication lists on three occasions in the month preceding the collection.
FUTURE DIRECTIONS
While the present study did not detect changes in the microbial composition over the course of 1 year, longer studies may be needed to assess if and how the GIT microbiome evolves over time in this disease state, particularly from the time of the initial diagnosis to the development of specific manifestations of SSc, such as ILD. Such studies may uncover novel microbial predictors of organ involvement in SSc. In addition, larger studies are necessary, which are adequately powered to detect important microbiota associations with specific SSc disease features.
CONCLUSION
This study demonstrated that both GIT symptoms and dysbiotic microbial composition remain relatively stable over the course of 1 year in patients with SSc. This study also identified specific bacterial genera associated with the SSc-GIT symptoms and was the first to do so in the context of a longitudinal study. Additional studies are needed to validate and expand upon these findings, particularly among patients with early SSc. Future studies may also assess whether manipulation of the GIT microbiome can lead to symptomatic improvement of SSc-GIT symptoms.