Abstract
Introduction: Prostate-specific antigen (PSA) is the main tool of detection for prostate cancer (PCa). However, PSA has limited specificity and sensitivity in determining the presence of PCa, leading to unnecessary biopsies and the diagnosis of potentially indolent PCa. The aim of this article is to review the tools available to urologists in the clinical situation of rising PSA with prior negative biopsies.
Evidence synthesis: The need for prostate biopsy is based on PSA level and/or a suspicious digital rectal examination. Ultrasound-guided biopsy is the current gold standard. The incidence of PCa detected by saturation repeat biopsy is 30–43%. Prostate health indes, prostate cancer antigen 3, and 4Kscore are available second-line tests to distinguish between malignant and benign prostate conditions, reducing the number of unnecessary biopsies. Molecular testing including ConfirmMDx (MDxHealth, Irvine, California, USA) and The Prostate Core Mitomic Test™ (PCMT) (MDNA Life Sciences, West Palm Beach, Florida, USA) are tissue tests for men with prior negative biopsy. Multiparametric magnetic resonance imaging (mpMRI) is used for lesion identification and subsequently for biopsy or treatment. In the setting of suspected PCa, the use of prostate mpMRI has shown to have a negative predictive value for clinically significant PCa of 80–96%.
Conclusions: Approximately 70% of patients undergoing prostate examination will have a negative result following analysis of the biopsy sample. This negative diagnosis leads to the common clinical challenge of determining when and if a repeat biopsy should be performed. New blood, urine, tissue, and imaging tools are now available to guide this decision.
INTRODUCTION
Prostate-specific antigen (PSA) testing is the main tool of detection for prostate cancer (PCa).1 However, PSA has limited specificity and sensitivity in determining the presence of PCa, leading to unnecessary biopsies and the diagnosis of potentially indolent PCa. Additional information may be gained by the Progensa DRE urine test (Hologic, Marlborough, Massachusetts, USA) for prostate cancer antigen 3 (PCA3), the serum 4Kscore and Prostate Health Index (PHI) test, or a tissue-based epigenetic test (ConfirmMDx). The current diagnostic procedure for men with suspected PCa is ultrasound-guided biopsy. For a prostate volume of 30–40 mL, >8 cores should be sampled; 10–12 core biopsies are recommended. Unlike many other solid tumours, for which image-guided biopsy is common, PCa has traditionally been detected by randomly sampling the entire organ. However, the recent introduction of multiparametric magnetic resonance imaging (mpMRI) allows for image-based identification, which may improve diagnostic accuracy for higher risk tumours. Advances in imaging have led to the development of fusion biopsy platforms in which mpMRI images are electronically superimposed in real time on transrectal ultrasound (TRUS) images. Numerous targeted biopsy platforms exist and can perform biopsies of suspicious regions seen on the prostate mpMRI.2-6 The aim of this article is to review the available tools for the urologist in the clinical situation of rising PSA with prior negative biopsies.
INDICATION FOR REPEAT PROSTATE BIOPSY
Current Role of Saturation Biopsies in Prostate Cancer
The need for prostate biopsy is typically based on PSA level and/or a suspicious digital rectal examination (DRE). Age, comorbidity, patient preference, and therapeutic consequences should also be considered and discussed beforehand. Risk stratification is a potential tool for reducing unnecessary biopsies. A single PSA elevation alone should not prompt immediate biopsy and the PSA level should be verified after a few weeks in the same laboratory. Empiric use of antibiotics in an asymptomatic patient to lower the PSA should not be undertaken.7,8
Ultrasound-guided biopsy is the current gold standard. The TRUS approach is used for most prostate biopsies although some urologists instead use a transperineal approach. PCa detection rates are comparable with both techniques according to two prospective randomised trials. Recently Scott et al.,9-11 in a retrospective cohort of 431 radical prostatectomy specimens, concluded that the transperineal approach predicted with more accuracy the clinical risk category than the TRUS approach. A higher level of evidence is needed to support these findings.
The actual indications for repeat biopsy include:
- Rising and/or persistently elevated PSA
- Suspicious DRE, 5–30% PCa risk
- Atypical small acinar proliferation, 40% risk
- Extensive (multiple biopsy sites, i.e. >3) high-grade prostatic intraepithelial neoplasia, 30% risk
- Positive mpMRI findings
The incidence of PCa detected by saturation repeat biopsy (>20 cores) is 30–43% and depends on the number of cores sampled during earlier biopsies. Saturation biopsy may be performed with the transperineal technique, which detects an additional 38% of PCa. The high rate of urinary retention (approximately 10%) is a drawback.12
Biomarkers (Table 1)
Screening, over-diagnosis, and over-treatment are topics of debate in PCa. There is a need to differentiate between clinically significant and indolent cancer, and PSA has been shown not to be the best marker to solve this issue. The ideal PCa biomarker would be capable of distinguishing PCa from benign prostate conditions and differentiating between aggressive and indolent tumours. Patients with a prior negative biopsy and persistently high PSA, especially in the grey area (4–10 ng/mL), represent a challenge and a controversial topic in which there is not currently consensus.13-15
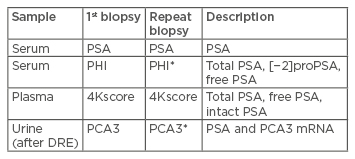
Table 1: Overview of biomarkers of prostate cancer.
*Approved by US Food and Drug Administration (FDA).
PSA: Prostate-specific antigen; PHI: Prostate Health Index; PCA3: prostate cancer antigen 3; DRE: digital rectal examination.
Accordingly, only 20–30% of men with serum PSA levels from 2–4 ng/mL and 30–45% with serum PSA levels from 4–10 ng/mL have PCa diagnosed on prostate needle biopsy. To address these limitations, adjunctive measurements, including the ratio of free-to-total PSA, called percent-free PSA, have been investigated and shown to significantly improve cancer detection rates within the 4–10 ng/mL range.16-18
More recently, distinct molecular forms of free PSA have been characterised and found to be differentially associated with benign prostatic hyperplasia (BPH) or PCa. These precursor forms of PSA are enzymatically inactive and include: i) proPSA, which is elevated in cancer tissue and serum, as well as ii) benign PSA (BPSA), and iii) intact PSA, which are associated with BPH. The [−2] proPSA isoform has emerged as a promising marker for PCa detection, as it is preferentially concentrated in cancerous tissue on histochemical staining.19
Prostate Health Index
The PHI blood test combines the relative concentrations of three different PSA forms: total PSA, free PSA, and [–2]proPSA, using a mathematical formula: ([–2]proPSA/free PSA) × √PSA. It was developed by Beckman Coulter in partnership with the National Cancer Institute (NCI) Early Detection Research Network (EDRN) and approved by the US Food and Drug Administration (FDA) in 2012. The 2016 National Comprehensive Cancer Network (NCCN) guidelines offer PHI as an option to increase specificity before initial or repeat biopsy, and has regulatory approval in >50 countries.20-22
PHI has been consistently shown to outperform PSA to distinguish malignant and benign prostate conditions in men with a PSA level >2 and/or a suspicious DRE. Several studies have demonstrated that PHI significantly improves PCa detection in high-risk cases and also predicts the aggressiveness of disease. In the clinic, PHI is less expensive than other tests such as the 4Kscore or PCA3, and does not require a physician to conduct a DRE, making it logistically attractive for both clinicians and patients.13
Recently, Loeb et al.23 developed a nomogram using continuous values of PHI as part of a multivariable model which improves the prediction of aggressive PCa among individual patients with PSA between 2–10 ng/mL and benign DRE. PHI predicted the risk of aggressive PCa across the spectrum of values. Adding PHI significantly improved the predictive accuracy of the risk calculators for aggressive disease. A new model was created using age, previous biopsy, prostate volume, PSA, and PHI, with an area under the curve (AUC) of 0.746. The bootstrap-corrected model showed good calibration with an observed risk for aggressive PCa and had net benefit on decision-curve analysis.23
Another recent study combining PHI and mpMRI in men requiring repeat biopsy explored the potential value of the PHI in the context of image-guided repeat biopsies. In this study, adding PHI to mpMRI improved overall and significant cancer prediction (AUC 0.71 and 0.75) compared to mpMRI + PSA alone (AUC 0.64 and 0.69, respectively). At a threshold of ≥35, PHI + mpMRI demonstrated a negative predictive value (NPV) of 0.97 for excluding significant tumours. In mpMRI negative men, the PHI again improved the prediction of significant cancers; AUC 0.76 versus 0.63 (mpMRI + PSA). Using a PHI ≥35, only 1/21 significant cancers was missed and 31/73 (42%) men were potentially spared a re-biopsy (NPV of 0.97, sensitivity 0.95). In this way, the authors proposed PHI adds predictive performance to image-guided detection of clinically significant cancers and has value in determining the need for re-biopsy in men with a negative mpMRI.24
Prostate cancer antigen 3
PCA3 was described initially by Bussemakers et al.25 in 1999. PCA3 score measures the ratio of PCA3 and PSA mRNA in the urine after vigorous DRE using transcription-mediated amplification.25,26 PCA3 was approved by the FDA in 2012 for men with a previous negative biopsy and a persistently elevated PSA level to aid in decision-making regarding repeat biopsies and was also an option mentioned in the 2016 NCCN guidelines. Although PCA3 can be offered to patients with a previous negative biopsy, its clinical effectiveness for this purpose is uncertain. In addition, its relationship to cancer aggressiveness is subject to debate and generally inferior to other markers.
Ferro et al.27 compared PHI with PCA3 in patients who were undergoing initial biopsy and found that PHI and PCA3 had a similar predictive accuracy for overall PCa detection; AUC of 0.77 for PHI and 0.73 for PCa and that both tests outperformed percentage-free PSA. In another study, PCA3 had similar predictive value for PCa in candidates for repeat biopsy compared to PHI (AUC 0.77 versus 0.69).27
In the repeat biopsy setting, there were also opposing results with no statistically significant differences between PHI and PCA3. Perdona et al.28 evaluated the use of a combination of PCA3 and PHI in predicting biopsy results in 160 men upon initial biopsy. Receiver operating characteristic (ROC) analyses showed that PHI outperformed PCA3 for high specificity level, whereas PCA3 outperformed PHI for high sensitivity level.28 On the other hand, Ferro et al.27 showed that PHI and PCA3 were the strongest predictors of PCa with no significant differences in pairwise comparison. The combination of the two tests did not further improve diagnostic power in this cohort.
Many studies show that PCA3 is inferior for identifying high-grade disease compared to PHI. Seisen et al.29 found that PCA3 detected more PCa overall than PHI when cut-off scores for positive results were set at >35 for PCA3 and >40 for PHI, but had worse performance than PHI for identifying clinically significant disease.
Ferro et al.30 found that PCA3 and PHI levels were significantly higher in patients with tumour volume ≥0.5 mL, pathological Gleason ≥7, and pT3 disease (all p values ≤0.01). ROC curve analysis showed that PHI is a better accurate predictor of high-stage (AUC 0.85 [0.77-0.93]), high-grade (AUC 0.83 [0.73-0.93]), and high-volume disease (AUC 0.94 [0.88-0.99]) than PCA3, who showed lower AUCs, ranging from 0.74 for Gleason to 0.86 for tumour volume.30
4Kscore
The 4Kscore is a risk calculator for the detection of PCa on biopsy, based on the 4-Kallikrein panel combined with patient age, DRE, and biopsy history. The 4-Kallikrein panel includes total PSA, free PSA, intact PSA, and hK2, a kallikrein with high homology with PSA responsible for the in vitro cleavage of proPSA, resulting in the ‘mature’ form of PSA. The 4Kscore provides probability of having high-risk PCa. Although the 4Kscore does not have FDA approval, the 2016 NCCN guidelines also offer this as a second-line testing option for patients who have never undergone biopsy or after a negative biopsy.22
The 4Kscore is associated with an improvement of 8–10% in predicting biopsy-confirmed PCa, indicating that the use of the 4Kscore could potentially reduce the number of prostate biopsies currently conducted by an estimated 48–56%.31
Tissue Markers (Table 2)
Sampling errors inherent with the random tissue collection of the biopsy procedure result in a false-negative rate of approximately 25%. This imprecision poses a diagnostic dilemma, often resulting in multiple repeat biopsies. Although diminishing rates of cancers are detected during these invasive repeat procedures, a high rate of clinically significant (i.e. a Gleason score ≥7) cancer is still on the second, third and fourth or more biopsies (65%, 53%, and 52%, respectively). Molecular testing is another option to help identify occult cancer in this situation.32-35
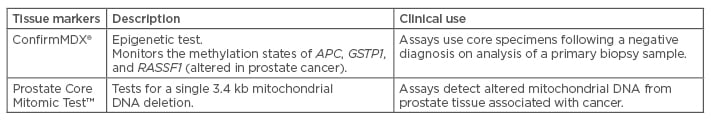
Table 2: Tissue markers of prostate cancer.
APC: adenomatous polyposis coli; GSTP1: glutathione Stransferase pi 1; RASSF1: Ras association domain family member 1.
ConfirmMDx
ConfirmMDx36 is a methylation assay that measures changes in methylation in benign tissue to identify peritumour regions adjacent to missed cancer (termed the ‘halo effect’). This test evaluates the methylation patterns of three genes: glutathione S transferase pi 1 (GSTP1), adenomatous polyposis coli (APC), and Ras association domain family member 1 (RASSF1). Investigators in the MATLOC study37 specifically examined the ConfirmMDx test by running this assay on core prostate biopsy samples from men with prior negative biopsy.36,37 After adjusting for patient characteristics, the assay was a significant predictor of repeat biopsy outcome on multivariate analysis (odds ratio [OR]: 3.17; 95% confidence interval [CI]: 1.81–5.53) with a NPV of 90%. A subsequent study of 350 American men demonstrated a NPV of 88% with ConfirmMDx the most significant independent predictor of PCa in repeat biopsy samples (OR: 2.69; 95% CI: 1.60–4.51).38
Of note, the presence of atypical features (such as atypical small acinar proliferation on histological examination) was also a significant predictor on multivariate analysis, associated with a two-fold increased risk of PCa diagnosis (OR: 2.11).38 Results to date suggest that this new tissue-based assay might help to distinguish between men who are free of PCa and those with occult disease, thereby potentially reducing the use of unnecessary repeat biopsy procedures. It is also included in the 2016 NCCN guidelines as an optional second-line test before repeat biopsy.
The Prostate Core Mitomic Test
PCMT39 is another field effect laboratory test based on detection of a single 3.4 kb mitochondrial DNA deletion. An early study involving a cohort of 183 men including those with benign, malignant, or pre-malignant biopsy samples, generated a remarkable AUC of 0.87 using this test in the validation phase.
In a follow-up study of 101 patients undergoing repeat biopsy procedures, 20 were found to have PCa within 1 year of the initial biopsy; analysis of biopsy samples for PCa using the PCMT was associated with a sensitivity and specificity of 84% and 54%, respectively, (AUC 0.75) and a negative predictive value of 91%.40 Larger validation studies are required before the widespread use of this assay can be recommended.
Problems with tissue-based assays include the potentially confounding effects of infection and/ or inflammation, age-related changes, the ability to distinguish high-grade from low-grade PCa, and the detection of clinically insignificant PCa. APC methylation patterns are altered in the presence of inflammation in many cancer types.41 Methylation patterns in histologically normal prostate tissue alter with age, potentially adding additional sources of error that must be considered. Overall, comparative studies are necessary to help guide test selection.
Multiparametric Magnetic Resonance Imaging
mpMRI of the prostate has been an imaging technique available for over two decades. In the past 10 years, there has been an increased interest in the method due to validation of multiple imaging parameters, which when combined can be used in PCa detection and staging.
mpMRI outputs are used by the radiologist for lesion identification or staging and subsequently can be used to perform targeted biopsy and to aid in treatment planning for those men ultimately diagnosed with PCa. The current European Association of Urology (EAU) recommendations outline its primary use in lesion targeting in the setting of a previous negative biopsy and a persistent clinical suspicion of cancer. Additionally, its use is recognised for local staging and in the decision process of whether to perform nerve-sparing surgery in the setting of intermediate or high-risk disease.1 A recent consensus statement from the American Urological Association (AUA) and Society of Abdominal Radiology (SAR) concluded that patients receiving a prostate imaging reporting and data system (PI-RADS) assessment category of 3–5 warrant repeat biopsy with image-guided targeting.
While TRUS-guided MRI fusion or in-bore MRI targeting may be valuable for more reliable targeting, in the absence of such targeting technologies, cognitive (visual) targeting remains a reasonable approach in skilled hands. At least two targeted cores should be obtained from each MRI-defined target. Given the number of studies showing a proportion of clinically significant cancers missed by MRI targeted cores, a case-specific decision must be made on whether to also perform concurrent systematic sampling. However, performing a solely targeted biopsy should only be considered once quality assurance efforts have validated the performance of prostate MRI interpretations with results consistent with the published literature. If a repeat biopsy is deferred based on MRI findings, then continued clinical and laboratory follow-up is advised and consideration should be given to incorporating repeat MRI in this diagnostic surveillance regimen.2
In the setting of suspected PCa and previous negative biopsy, the use of prostate mpMRI has shown to have a NPV for a clinically significant PCa of 80–96%.3-6 Its use in this cohort is primarily by planning a mpMRI targeted biopsy, which has been demonstrated to detect more clinically significant cancers and fewer clinically insignificant cancers than a systematic biopsy.6 The best fusion biopsy method and approach is still strongly debated, with vendors offering either a TRUS or transperineal biopsy approach. That notwithstanding, fusion biopsy appears to improve sampling efficiency for clinically significant disease regardless of the platform.2,42
Image reporting has been standardised in 2012 with the PI-RADS classification, later revised in 2015 as Version 2.43,44 Image acquisition is recommended to be performed on a high-field magnet (≥1.5 tesla), with or without the use of an endorectal coil.44 Interpretation is performed based on three imaging sequences: T2-weighted imaging, diffusion-weighted imaging (DWI), and dynamic contrast enhancement (DCE).
T2-weighted imaging is used to delineate prostate anatomy, identify suspicious areas of clinically significant PCa, and assess extracapsular extension.1 Current PI-RADS recommendations consider it the dominant sequence in lesion scoring in the transition zone of the prostate, with about 30% of PCa occurring in this region.45 In the peripheral zone, PCa is seen as a mass-like low signal lesion, but this appearance has a low specificity and is considered insufficient on its own, which led to the addition of DWI.46
DWI allows for the measurement of water movement through a tissue voxel. A high signal indicates slower water diffusion through tissue indicating a high cell count or tissue swelling. Due to the higher mitotic rates of neoplastic cells, areas with PCa cells may be visible as bright areas (i.e. areas of restricted diffusion). This is useful in the homogenous peripheral zone of the prostate, where >70% of cancers are detected.45 For this reason, the dominant sequence for assessment of the peripheral zone is DWI. This sequence is technically difficult to obtain well, being dependent on multiple factors, which has led to the addition of DCE imaging to the diagnostic protocol.
Contrast administration is acquired as a dynamic sequence, allowing for the registration of temporal contrast curves. The reviewed PI-RADS guidelines have decreased the diagnostic importance of focal enhancement. Currently, DCE is used to further characterise lesions of undetermined significance in the transitional zone (PI-RADS 3). The additional finding of enhancement of these lesions increases the lesion probability to PI-RADS 4, and its influence on scoring is binary. The use of gadolinium is currently debated, and may be of less importance in future guidelines.47,48
Prostate mpMRI has emerged as an effective diagnostic tool used in PCa detection and subsequently may help with staging for those men ultimately diagnosed with PCa. Its use is facilitating a shift from the traditional PSA-TRUS biopsy pathway to a patient-centred model according to the tumour location and stage, making its applications invaluable.
CONCLUSIONS
Approximately 70% of patients who undergo prostate biopsy will have a negative result. This negative diagnosis leads to the common clinical challenge of determining when and if a repeat biopsy should be performed, and which tools should be used to guide this decision. Despite all the current evidence, no recommendation can be made regarding the best biomarker to use in the setting of repeat biopsies; although PHI, 4Kscore, and PCA3 have added value in the detection of PCa on biopsy. The use of new molecular diagnostic technologies, such as epigenetic tests, is another possible way to inform repeat biopsy decisions. Meanwhile, comparative studies are needed to confirm the optimal biomarker to determine which is the most cost-effective marker to use in patient selection for repeat biopsy.
Biomarkers in PCa are a rapidly expanding field and recent developments of proteomic/genomic platforms, as well as the rise of immunotherapy provide meaningful research opportunities for the upcoming years. Other promising innovations, such as imaging biomarkers, are also being developed. Nonetheless, many challenges still lie ahead. In the current clinical practice, before repeat biopsy, the urologist should perform mpMRI when clinical suspicion of PCa persists in spite of negative biopsies, with a 1a level of evidence and grade of recommendation A. A major goal to strive for is refinement of the current biopsy approach by using mpMRI in optimising the detection of significant PCa.