Meeting Summary
Cancer of unknown primary (CUP) accounts for 3–5% of all cancers,3,4 and prognosis is poor for most patients, with a median survival of 6–9 months.5 Clinical and pathological diagnostic work-up is required to determine whether patients belong to the favourable or unfavourable subset of CUP. Only 15–20% of patients belong to favourable subsets and have responses to therapy and outcomes similar to those of patients with the equivalent known metastatic primary tumour.4
For the patients in the unfavourable subsets (around 80–85% of CUP patients) treatment to date has been with chemotherapy. Median survival is <1 year5 and clinicopathological management of these patients is not expected to improve outcomes further. However, two different approaches involving genetic testing to guide patient management have the potential to offer progress.
The first approach is to use gene or methylation profiling tests to identify the tissue of origin. A number of tests are available that can be used to examine the gene expression or methylation signature of the CUP sample and assign a tissue of origin biologically. This approach is being used in clinical trials,6 but there is not yet solid clinical evidence that offering primary-specific therapy to these patients improves outcomes.
The second approach is to identify genomic aberrations that can be targeted therapeutically. Comprehensive genomic profiling (CGP) can identify aberrations that can be targeted with available agents in some patients,1 but there is no high-level evidence concluding that this approach improves outcomes. A novel molecularly guided trial, CUPISCO,7 was recently initiated and will address this issue in a Phase II, randomised, active-controlled, multicentre setting in patients with newly diagnosed, poor-prognosis CUP. The study aims to show the benefit associated with the use of genomic profiling to allocate molecularly targeted therapies or immunotherapies compared with the standard treatment of platinum-based chemotherapy in patients with CUP.
Introduction
CUP is a heterogeneous disease with diverse origins, meaning the primary tumour has not been identified. CUP is essentially a disease of metastases that disseminate early and aggressively, and for which standard clinical and pathological diagnostic work-up does not identify the primary tumour. Multiple metastases develop in an unpredictable pattern.5
The disease is not as rare as is sometimes assumed; it accounts for 3–5% of all cancers3,4 and is the fourth most common cause of cancer-related deaths.8 Prognosis is poor in the vast majority of patients. The median survival is 6–9 months5 and only 15–20% of patients belong to a favourable subset with a median overall survival of 12–36 months.9 There is a high unmet need for new treatments for CUP, and its management remains a major challenge.
Cancers of Unknown Primary: Do Clinical and Standard Pathological Decision Parameters Dominate the Treatment Algorithms or Can Treatment be Improved Using Genetic Profiling?
Professor George Pentheroudakis
An Overview of Clinical and Pathological Management of Cancer of Unknown Primary Patients
The standardised diagnostic work-up is defined in the European Society for Medical Oncology (ESMO) Clinical Practice Guidelines.9 The first step involves thorough pathological and clinical diagnostic work-up. The pathological diagnosis involves immunohistochemistry of tissue samples to rule out treatable tumours, such as germ cell cancers, lymphomas, melanomas, and sarcomas. Once the tumour is known to be epithelial, cytokeratin (CK) cocktails, such as CK7/CK20, allow the pathologist to assign a tissue of suggestive origin to the CUP tumour. Although in most cases the pathologist can determine whether the CUP is of epithelial origin, frequently it is not possible to suggest the specific epithelial tissue of origin of the tumour with a high level of confidence.
The clinical work-up comprises a thorough medical history, physical examination, and CT scans of the head, thorax, abdomen, and pelvis. Female patients should have a mammography. In most CUP cases, several tumour markers are elevated in the blood, but this often does not help determine the primary tissue of origin. The ESMO Guidelines9 suggest testing for serum α-fetoprotein and human chorionic gonadotropin in patients with midline metastatic nodal disease and in patients with liver metastases in order to not miss hepatoma and germ cell tumours. A serum prostate-specific antigen test is recommended for males with adenocarcinomatous bony metastases. Signs, symptoms, and laboratory abnormalities direct the physician towards additional relevant procedures. Head and neck PET and CT scans are only useful if locoregional therapy is being considered for the CUP patient, for example, in patients with squamous cell carcinoma affecting the cervical lymph nodes.
A key decision-making point in the management of CUP is to decide whether the CUP patient belongs to favourable or unfavourable subsets; only 15–20% of CUP patients belong to favourable subsets.9
Favourable subsets include men with poorly differentiated carcinoma with midline nodal distribution (extragonadal germ cell syndrome); women with papillary serous adenocarcinoma of the peritoneal cavity; women with adenocarcinoma involving only axillary lymph nodes, squamous cell carcinoma involving cervical lymph nodes, or poorly differentiated neuroendocrine carcinomas; men with blastic bone metastases and/or elevated prostate-specific antigen; patients with metastases with a colon cancer immunohistochemical profile (CK7-, CK20+, CDX2+); and patients with single-site metastases.4
Cancers in these subsets are very likely to respond to primary-specific treatment, which translates into a better prognosis compared to patients belonging to the large group with unfavourable prognosis CUP. Prof Pentheroudakis argued that, from a biological standpoint, these tumours are not CUP in a strict sense. Several retrospective series examining the natural behaviour and biology of CUP tumours in favourable subset patients have suggested patients in this group have responses to therapy and outcomes that are similar to those with the equivalent known metastatic primary tumour.4 For example, women with axillary lymph node adenocarcinoma can be managed as if they have breast cancer, and patients with squamous cell carcinoma in the cervical lymph nodes can be treated as if they have head and neck cancer.
The unfavourable subsets encompass 80% of patients with CUP. These patients typically have several metastases in the viscera (liver, bones, veins, lungs) and no identifiable primary tumour. To date, treatment has been platinum-based chemotherapy and median survival is <1 year. Prof Pentheroudakis commented that the clinicopathological approach to treatment of CUP will not improve outcomes further.
Patient Management Relies on Two Distinct Approaches
Genetic testing has the potential to offer new lines of progress. There are two main approaches: the first is to use molecular tests to identify the tissue of origin, while the second is to use tests to find genomic aberrations that can be targeted therapeutically.
The first approach, identifying the tissue of origin, rests on the premise that each tumour type has a distinct molecular profile, identified by examination of RNA expression or gene methylation analysis of the CUP sample. The expression signature is compared to known expression signatures from several solid tumours and a CUP sample is assigned a tissue of origin biologically; this was not possible by clinicopathological means. These tests have been validated in typical metastatic solid tumours and were shown to have an accuracy of 85–90%.10 A number of tests are available, which analyse between 10 and 2,000 genes simultaneously and distinguish between 6 and 50 different cancer types. They variously use messenger RNA, microRNA, or DNA methylation.
To date, there is no evidence of improved outcomes using molecular testing that enables the patient with CUP to receive primary-specific therapy. The best evidence comes from a cohort study led by investigators at the Sarah Cannon Cancer Institute in Dallas, Texas, USA.11 Of 289 treatment-naïve CUP patients enrolled, assays were taken successfully in 252 patients and 247 (98%) had a tissue of origin predicted. Primary-specific therapy was received by 194 patients and in this group, median survival was 12.5 months (95% confidence interval: 9.1–15.4 months). Prof Pentheroudakis said this is slightly longer than the overall survival rates historically seen with chemotherapy, but the difference is small. He also noted that the trial was not randomised.
An ongoing randomised French trial, GEFCAPI 04,6 including 223 treatment-naïve patients with CUP, may provide clearer answers. Molecular profiling of tissue samples from all patients has been carried out with the aim of assigning biologically a tissue of origin. Patients have been randomised into blinded or unblinded groups; in the unblinded arm of the trial, the results of profiling were known to the investigator and the patient received primary-specific therapy. In the blinded arm, results were not disclosed and patients were treated with standard chemotherapy with cisplatin and gemcitabine. The primary endpoint is progression-free survival and the results are expected in 2019.
The second approach uses genomic profiling to find genomic aberrations that can be targeted therapeutically. The two most commonly cited tests are the Foundation One12 and Caris Life Sciences13 assays. In 2015, the Foundation One platform was used to perform a mutation analysis of 236 genes along with next-generation sequencing of rearrangements in 19 genes. One study found that the test identified mutations that could be targeted with currently available drugs in 20% of cases (n=200).14 In 2014, the Caris Life Sciences platform was used to perform a mutation analysis of 47 genes, immunohistochemistry for 23 markers, and fluorescence/chromogenic in situ hybridisation of 7 genes. It was used to profile 1,806 CUP cases and biologically relevant mutations were found in almost all cases (96%).15 However, 3 years later, investigators used an updated version of the Caris platform, a 592 gene NextSeq panel, to profile 389 CUP cases and identified therapeutically targetable mutations in 22% of patients.16 The latter result is more realistic, Prof Pentheroudakis concluded.
If genomic profiling identifies a targetable mutation in a tissue sample of CUP, and the patient receives targeted treatment, is the outcome improved as compared to standard chemotherapy? This approach has been trialled in metastatic solid tumours,17 but not in CUP, Prof Pentheroudakis said. The question of whether this approach is effective in CUP will not be answered before results are available from trials such as CUPISCO (Figure 1).7 This randomised Phase II trial will compare platinum-based chemotherapy with molecularly-targeted therapies relevant for the aberrations found by genomic profiling. This study includes only patients with histologically confirmed CUP and Prof Pentheroudakis commented it is expected to provide hints as to whether the approach is effective.
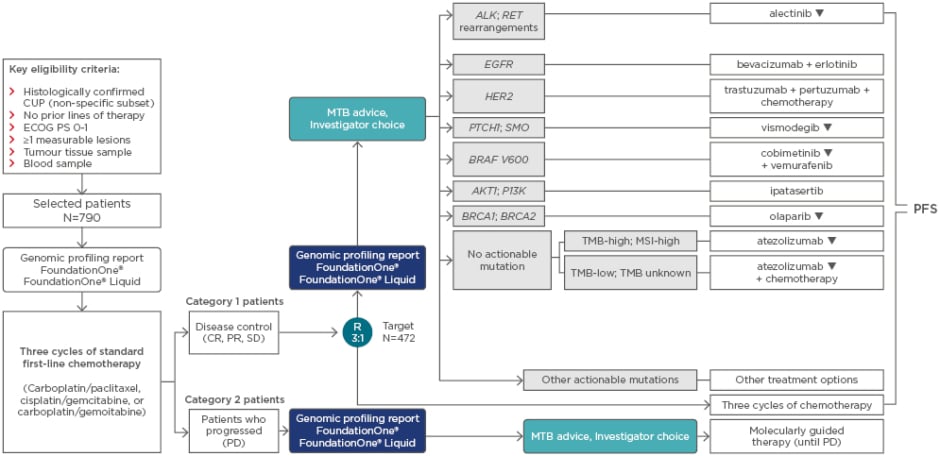
Figure 1: CUPISCO study design.
Randomisation is stratified by sex and response during the induction period (CR and PR versus SD). Genes listed comprise the confirmed selection of variants. Experimental genes may also be used for therapy selection. Therapies marked with ▼ are subject to additional monitoring. Reporting suspected adverse reactions after authorisation of the medicinal product is important and adverse events should be reported to your respective local office.
CR: complete response; CUP: cancer of unknown primary; ECOG PS: Eastern Cooperative Oncology Group performance status; MSI: microsatellite instability; MTB: Molecular Tumour Board; PD: progressive disease; PFS: progression-free survival; PR: partial response; R: randomisation; SD: stable disease; TMB: tumour mutational burden.
Adapted from Krämer et al.18
Overall, Prof Pentheroudakis stated that there is not yet high-level evidence to suggest that treating CUP patients with primary tumour-specific therapy, or targeting relevant genomic aberrations, does indeed improve outcomes for these patients. Trials such as GEFCAPI 046 and CUPISCO7 will provide long-awaited answers.
A Proposal for Patient Management in Cancers of Unknown Primary
Prof Pentheroudakis outlined a scheme for the management of CUP patients. When a patient presents with possible CUP, the first stage of management is clinical evaluation and biopsy. Anamnesis, physical examination, imaging, and standard pathology investigations using immunohistochemistry can identify the primary site, in which case the patient does not have CUP.
If the primary site is not identified by imaging or pathological diagnosis, the next step uses clinicopathological information to determine whether the patient belongs to one of the favourable subsets of CUP. Patients belonging to these subsets are treated with primary- specific therapy. However, 80–85% of CUP patients have non-favourable CUP. In theory, gene expression and/or gene methylation profiling could assign the tissue of origin in 80–85% of these cases,10 and the choice then is either to offer primary-specific therapy or enter the patient into a clinical trial.
In the remaining 15–20% of cases, the tissue of origin is not identified, despite profiling analyses,10 because of a failure of the test or an inadequate sample. The option for these patients is either a clinical trial or next- generation sequencing profiling of the tumour to find a targetable mutation. If a genomic aberration is found, treatment still depends on access to targeted drugs. Prof Pentheroudakis said that in countries such as Greece, access to targeted drugs is difficult because their use in CUP is not included in the approved indications. Furthermore, he stated that there is no high-level evidence to guarantee that this targeted approach will be effective. For example, a BRAF mutation may, in theory, be effectively treated if the CUP has metastasised from melanoma, but not if it has metastasised from colon cancer. Data from clinical trials are necessary to resolve this question.
In conclusion, it is first important to decide whether patients have favourable versus unfavourable CUP. Molecular testing may be able to assign the tissue of origin and to identify targetable genomic aberrations. However, high-level evidence to prove that targeting specific mutations will improve patient outcomes is still missing; ongoing clinical trials will address this question.
Addressing Unmet Needs in Cancers of Unknown Primary with a Novel Molecularly-Guided Trial
Professor Alwin Krämer
At ESMO 2018, Prof Krämer also highlighted the importance of genomic profiling in this setting. At a sponsored symposium entitled ‘Comprehensive genomic profiling: Taking precision medicine from vision to reality’, he reviewed the evidence supporting the use of genomic profiling in patients with CUP.
In 2015, a study also highlighted by Prof Pentheroudakis profiled CUP samples using Foundation One CGP; 125 patients had adenocarcinoma of unknown primary and 75 had non-adeno-CUP syndrome. Of a total of 200 CUP cases profiled, 96% harboured at least one genomic alteration and ≥1 clinically relevant genomic alteration was identified in 85% of cases (169 out of 200).14
The types of genomic alterations identified showed a pattern similar to those seen in many other cancer types: few genes had frequent aberrations and vast numbers of other genes had rare aberrations, including some that would be amenable to targeted treatment.
Furthermore, a retrospective analysis of 6,116 CUP samples from the Foundation Core database identified complex immune genomic signatures using CGP. This study found that approximately 10% of patients with CUP harboured a high tumour mutational burden (TMB) status.19 Another analysis of 4,210 samples found that 1.6% showed a high microsatellite instability (MSI) status.19 These genomic signatures broaden treatment options to include immune-oncology therapies.
Earlier this year, Prof Krämer presented research at the American Society of Clinical Oncology (ASCO) Annual Meeting 2018, examining the overlap between different pathways in a population of CUP patients1 in collaboration with Foundation Medicine.
The objective of the study was to analyse genomic profiles of CUP samples and characterise the association between clinical phenotype, affected signalling pathways, and candidacy for molecularly targeted therapies or immune checkpoint inhibitor (ICPI) treatment. Biopsies from 4,650 patients with CUP were sequenced with FoundationOne CGP and actionable alterations were identified for the majority of patients. A median of three genomic aberrations per sample was identified. Most samples (3,675 out of 4,650; 79.0%) harboured ≥1 genomic aberration or biomarker relevant for targeted therapy or ICPI; an additional 485 (10%) had genomic aberrations associated only with an investigational targeted therapy. More than one-third of the group (1,767 out of 4,650; 38%) harboured a genomic aberration specific for one of eight common targeted therapy/ICPI strategies, with 275 (5.9%) having a profile relevant to >1 of those eight strategies.
Profiling also offered insights into CUP subtypes that may be associated with reduced efficacy for some therapies: of 554 (12.1%) samples with high TMB or MSI, 145 (26.7%) harboured a genomic aberration known or suspected to reduce ICPI sensitivity (Table 1).
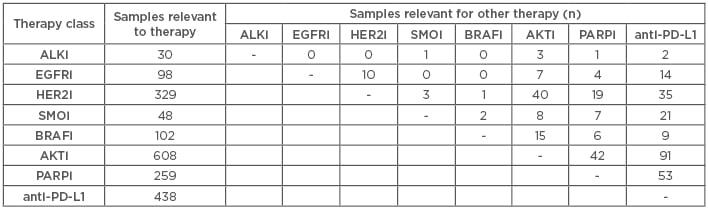
Table 1: Genomic profiling of carcinomas of unknown primary to support clinical decisions.
anti-PD-L1: anti-programmed death-ligand 1; i: inhibitor.
The study concluded that this knowledge-based approach in CUP patients describes informative genomic features. Coupled with the availability of a growing collection of targeted agents and immunotherapies, a new and rationally designed treatment paradigm, independent of tissue of origin, may now be possible in CUP. This approach can be assessed in prospective randomised trials investigating targeted therapies and ICPI; however, clinical studies evaluating this potentially promising approach are currently lacking.
At the ESMO Congress, Prof Krämer said these data were confirmed by another study on genomic aberrations in CUP, which was performed at the Memorial Sloan Kettering Cancer Center (MSKCC), New York City, New York, USA.20 Investigators used MSK-IMPACT technology, a deep coverage hybridisation capture-based assay encompassing 341 cancer-associated genes (later expanded to 410 cancer-associated genes). In this study, 333 CUP patients were evaluated and profiling was performed on samples from 150 patients. The results showed that 30% of cases (n=45 out of 150) harboured ≥1 lesion that was amenable to targeted therapy by licensed drugs. Of this group, 10% (15 out of 150) received matched therapy. Time to treatment failure ranged from <1–14 months, with several patients remaining on targeted therapy at the time of data cut-off. An additional 14% of patients had dominant mutation signatures, including high TMB and high MSI,20 that, again, suggest that immune-oncology therapies would be appropriate.
Together, these results suggest that genomic profiling can identify clinically relevant genomic alterations and direct new treatment options in patients with CUP. However, there remains a lack of high-level evidence to demonstrate the efficacy and safety of this approach. Clinical trials are needed to compare molecularly guided therapy versus standard chemotherapy across a large cohort of patients with CUP.
CUPISCO: Comprehensive Genomic Profiling and Molecularly-Guided Therapy in Cancer of Unknown Primary
The aforementioned data form the rationale for the CUPISCO trial, recently initiated by Roche and presented at the ESMO Congress 2018.2 CUPISCO is a Phase II, randomised, active-controlled, multicentre study of patients with newly diagnosed, unfavourable CUP.7 The study will compare the efficacy and safety of targeted therapy or immunotherapy guided by genomic profiling with platinum-based standard chemotherapy.
The trial will be launched in 101 institutions in 23 countries and will include 790 patients with CUP; it has been activated in 15 of the 23 countries already. Adults with newly diagnosed, poor prognosis CUP, as described in the ESMO Guidelines,9 are eligible to enter the trial. Exclusion criteria include non-epithelial cancer, squamous-cell CUP, and patients belonging to favourable subsets of CUP.
As outlined in Figure 1, all enrolled patients receive genomic profiling with Foundation Medicine Tumor Profiling of tissue and blood; all also receive three induction cycles of platinum-based chemotherapy. Those who respond to these three initial cycles of chemotherapy are randomised in a 3:1 ratio either to experimental treatment (n=354) or to the standard treatment arm (n=118) of continued chemotherapy for an additional three cycles. The experimental treatment arm is composed of nine strata depending on the alteration identified: seven of the strata are molecularly-targeted therapies to an identified genomic aberration, while an additional two arms cover immunotherapy. Patients with TMB-high or MSI-high tumours receive atezolizumab alone, while those with TMB-low or unknown receive atezolizumab in combination with platinum-based chemotherapy.
Patients whose tumours harbour a mutation that could be targeted by a specific drug not provided in the experimental study arm can receive this treatment in a tenth stratum of the study. Those whose disease progresses after the first three induction cycles of chemotherapy will have access to the targeted treatments shown in the experimental arm of the trial but in a non-randomised fashion.
A key element of the study design is a molecular tumour board, comprising the investigator, reference pathologist, reference oncologist, and, when appropriate, a cancer genomics consultant. The board will advise on experimental therapy choice for patients randomised to molecularly-guided therapy and for those who did not respond to the induction chemotherapy. As highlighted in the aforementioned ASCO abstract,1 given the overlap of some alterations, the choice of targeted therapy may be ambiguous in some cases (approximately 6%), but the molecular tumour board will follow a charter of guidance for therapy selection in hese cases.
In patients who responded to induction chemotherapy, the primary endpoint in CUPISCO is progression-free survival, defined as the time from randomisation to the first occurrence of disease progression or death from any cause. Secondary endpoints are overall survival, objective response rate, and duration of clinical benefit.
Response will be assessed by the investigator via a physical examination, CT scans, and MRI, using Response Evaluation Criteria In Solid Tumors (RECIST version 1.1) at the end of the induction period, every 3 treatment cycles and every 3 months during follow-up. Adverse events (AE) will be monitored and documented continuously during the study, and serious AE will also be documented and reported, as will AE of special interest.
The prospective randomised CUPISCO study has been set up against a background of a lack of high-level evidence to support potentially promising new approaches in the treatment of CUP. With the advent of large-scale DNA sequencing technologies, and the availability of a growing collection of targeted agents and immunotherapies, a new and rationally designed treatment paradigm may now be possible for CUP that is independent of the tumour of origin and customised to the patient.
Conclusion
CUP is associated with a poor prognosis and represents a high unmet medical need. For patients presenting with CUP, thorough clinical and pathological diagnostic work-up is necessary to establish whether the disease belongs to the favourable or unfavourable subsets. For patients whose disease falls into the unfavourable subsets, new molecular tests offer the possibility of assigning biologically a tissue of origin. When this is not possible, genomic profiling may identify genomic aberrations that can be targeted with available drugs; it has been established that most CUP carry at least one such aberration. Furthermore, CUP represents an ideal model to test the clinical utility of genomic profiling in a histology-independent setting. It allows for a pan-cancer analysis of the function and effect of targeted therapies and immunotherapy in an, until recently, almost neglected disease.
However, to date there is a lack of high-level evidence to suggest that genetic approaches improve the outcomes of CUP patients. CUPISCO is a novel clinical trial that aims to show benefit associated with genomic profiling used to define molecularly-targeted treatments. The Phase II trial of patients with newly diagnosed unfavourable CUP will compare the safety and efficacy of these targeted therapies with standard platinum-based chemotherapy. Clinical trials such as CUPISCO are necessary before a new and rationally designed treatment paradigm will become standard management for patients with CUP.








